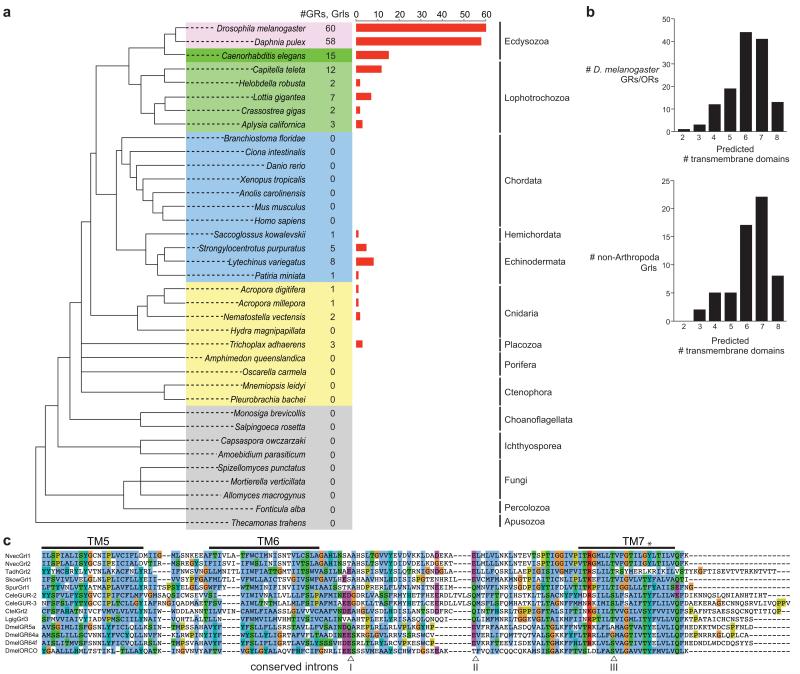
Figure 1. Identification of Gr-like (Grl) genes.
(a) Summary of the GR/Grl repertoires identified in the genomes of selected arthropods (pink), non-arthropod Ecdysozoa (dark green), Lophotrochozoa (light green), Deuterostomia (blue) and non-Bilateria (yellow). An unscaled tree showing the phylogenetic relationships between these species is illustrated on the left; relationships among the different non-bilaterian phyla is unresolved, with the exception of the Cnidaria, which are the closest sister group to Bilateria62. Related genes were identified in multiple species of nematode worms but, for simplicity, only C. elegans is shown.
(b) Computational predictions of the number of transmembrane domains found in D. melanogaster GRs and ORs (top) and Grls (bottom). Grl sequence fragments (lacking start/stop codons) were excluded from this analysis.
(c) Alignment of the C-terminal regions of select insect GRs and ORs, C. elegans GURs and Grls. Predicted transmembrane (TM) domains are indicated with horizontal lines. The asterisk marks a conserved tyrosine residue important for ion conduction in insect ORs31. The arrowheads below the alignment indicate the positions of three phase 0 ancestral GR introns (inferred from analysis of D. melanogaster GRs and ORs1) that are conserved in Grls: intron I is conserved in phase and position in NvecGrl1, NvecGrl2, TadhGrl2, SpurGrl1, CeleGUR2, CeleGUR3, CtelGrl2, LgigGrl3; intron II is conserved in phase and position in NvecGrl1, NvecGrl2, TadhGrl2, SpurGrl1, CtelGrl2, and LgigGrl3. Intron III is conserved in all sequences.