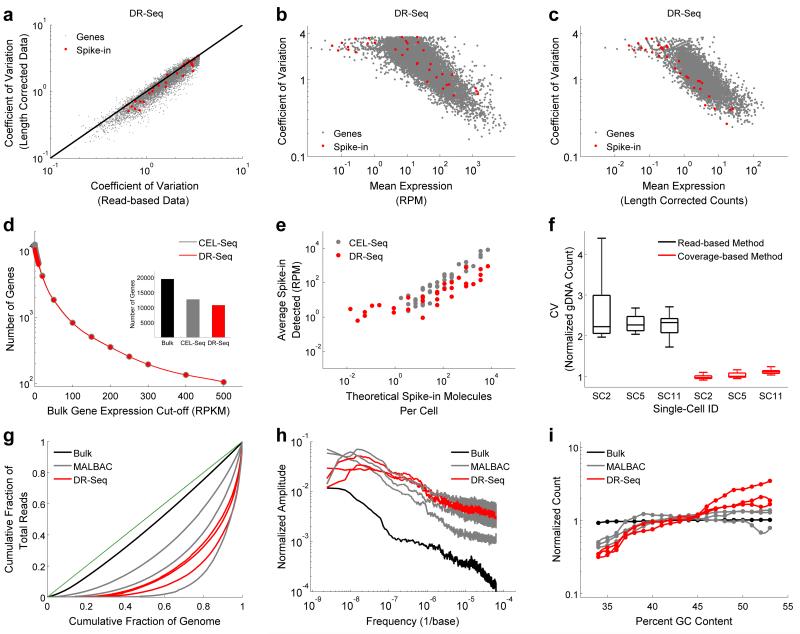
Figure 2.
Development of a computational techniques to reduce technical noise in single-cell DR-Seq sequencing data and comparison of DR-Seq to existing single-cell gDNA or mRNA sequencing methods in the mouse embryonic stem cell line E14. (a) Comparison of the coefficient of variation showed that cell-to-cell variability in the expression of genes reduced after correcting the raw read-based data using length-based identifiers, implying reduction in technical noise in the single-cell transcriptome data of DR-Seq (also see Supplementary Fig. 6). (b) Coefficient of variation versus mean expression of genes for the read-based data. Because each cell contains the same number of spike-in molecules, they are expected to display the lowest noise for a given mean level of expression. The data shows that read-based data contains significant amount of technical noise that obscures biological variability between single cells. (c) After correcting the DR-Seq data using length-based identifiers, spike-in molecules typically display the least noise over the entire range of mean expressions (also see Supplementary Fig. 7). Endogenous genes and spike-in molecules are indicated using gray and red dots, respectively. (d) Comparison of mRNA sequencing results between DR-Seq and CEL-Seq showed that both methods show similar performance in detecting genes above different expression thresholds obtained from bulk mRNA sequencing data. (Inset) Overall, both methods detect similar number of genes (also see Supplementary Fig. 10). (e) Detection of ERCC spike-in molecules in both methods increased monotonically with the expected number of molecules per cell. The figure shows spike-ins that were found in at least 2 single cells. (f) Box plot comparing bin-to-bin variability in gDNA read counts using two different methods for 3 single cells amplified by DR-Seq. The coverage-based method displays approximately two-fold reduction in technical noise compared to the read-based method. The box plots show the coefficient of variation of read distribution over all the autosomes in the mouse genome. (g) Lorenz plots were used to compare single cell gDNA sequencing results between DR-Seq and MALBAC. Lorenz curves were used to assess the uniformity of genome coverage by plotting the cumulative increase in read depth verses the cumulative fraction of genome covered, ordered by increasing coverage. The green line indicates the theoretical limit with reads distributed uniformly across the whole genome. Based on the Lorenz plots, bulk sequencing achieves read distribution close to the theoretical limit. The 6 single cells processed with either DR-Seq or MALBAC display similar distribution of reads across the genome. (h) Power spectrum of read distribution over different genomic length scales are shown for bulk sequencing and single cells processed by DR-Seq and MALBAC. The power spectrum reveals biases in read depth distribution over different ranges of genomic length scales. Bulk sequencing shows the least bias in read distribution with both DR-Seq and MALBAC performing similarly. (i) Read distribution for regions of the genome with different GC content shows that both methods deviate from the expected normalized count of 1 for regions with high and low GC content. This GC bias is corrected prior to estimating copy numbers in single cells.