Figure 4.
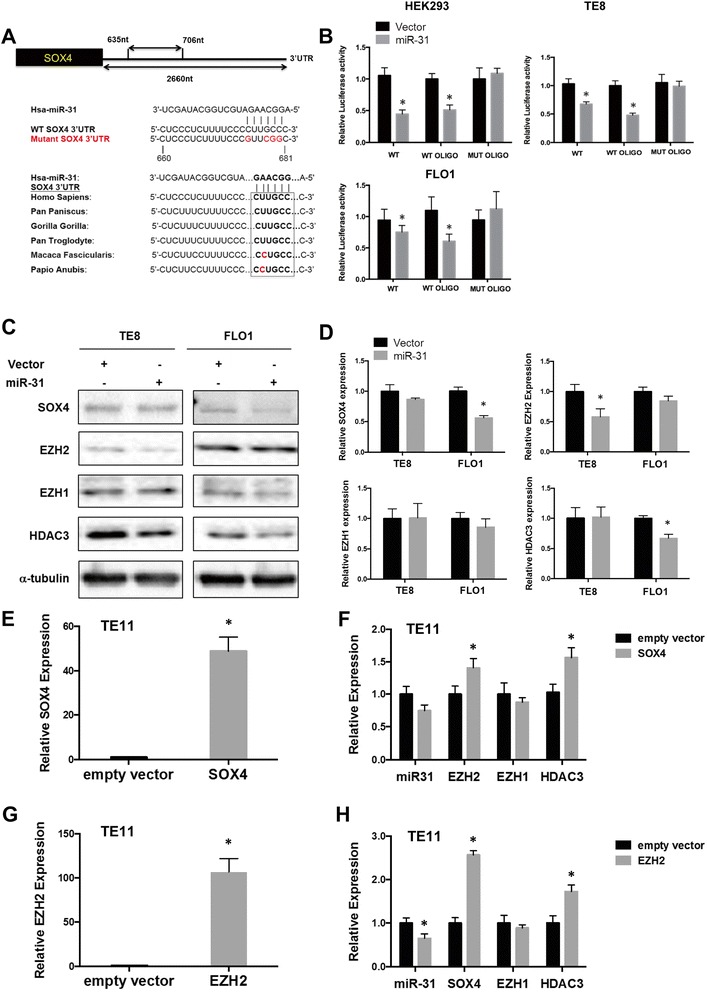
miR-31 directly targets SOX4 and indirectly targets EZH2 and HDAC3. (A) Computational analysis revealed one miR-31 binding site in the 3′UTR of SOX4. The upper panel shows the region containing the miR-31 binding site. The mutated SOX4 3′UTR seed region is indicated. A SOX4 3′UTR fragment containing wild type (WT OLIGO) or mutant (MUT OLIGO) of the miR-31-binding sequence was cloned into the downstream of the luciferase reporter gene. The lower panel shows the nucleotide sequence alignment of the predicted miR-31 binding site in the 3′UTR of SOX4 of six species. (B) HEK293, TE8 and FLO1 cells were co-transfected with psiCHECK-2 dual Renilla/Firefly luciferase plasmid containing either wild-type, wild-type oligo or mutant oligo of SOX4 3′UTR (indicated as WT, WT OLIGO and MUT OLIGO) with either pBABE empty vector control or pBABE-miR-31 vector. Luciferase activity was determined 48 hrs after transfection. (C) TE8 and FLO1 cells were transfected with miR-31 vector or empty vector control and cell lysates were analyzed after 72 hrs for SOX4, EZH2, EZH1 and HDAC3 by western blotting. α-tubulin was used as an internal control. (D) qRT-PCR analysis of SOX4, EZH2, EZH1 and HDAC expression in TE8 and FLO1 cells transfected with miR-31 or empty vector control. (E, F) qRT-PCR analysis of TE11 cells transfected with SOX4 or empty vector control. (G, H) qRT-PCR analysis of TE11 cells transfected with EZH2 or empty vector control. miR-31 expression was normalized to RNU6 and SOX4, EZH2, EZH1 and HDAC3 were normalized to GAPDH. Results are means ± SD from at least three biological replicates.
