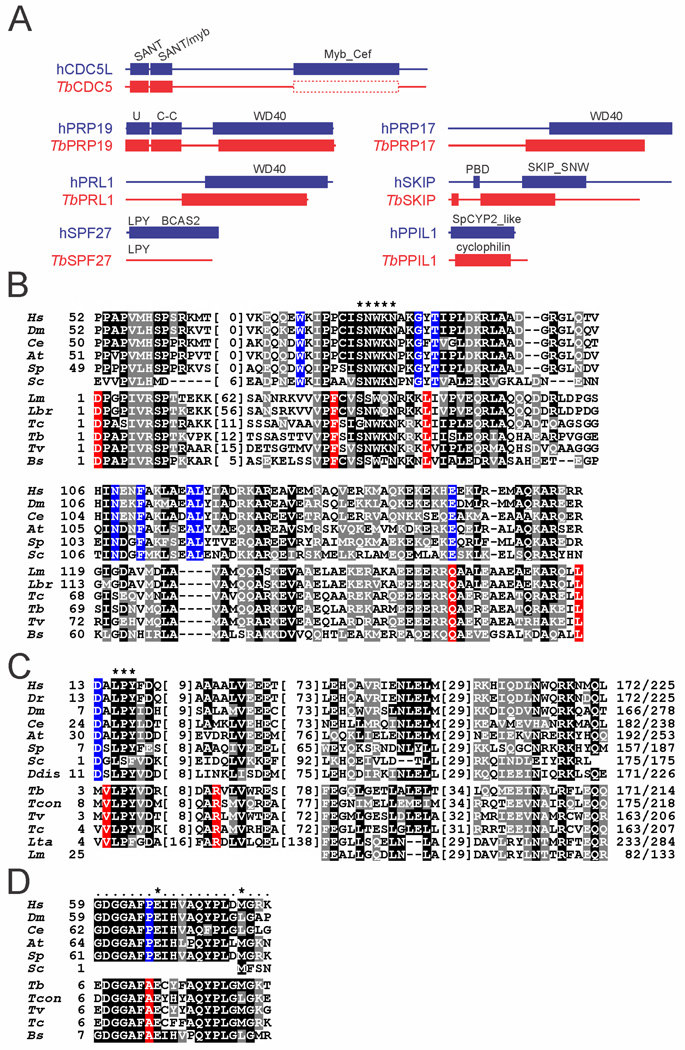
Fig. 2. Domain structure and sequence conservation in PRP19 complex subunits.
A. Domain structure drawn to scale of trypanosome PRP19 complex subunits and their human orthologs. The protein domains were identified by NCBI blastp searches of the Conserved Domain Database (Marchler-Bauer et al., 2011) (E-values are specified for T. brucei protein domains): CDC5: SANT domain (cd00167), E = 1.03e−08; SANT/myb-like domain (cd11659), E = 8.07e−10; the very limited sequence similarity between the human Myb_Cef domain (pfam11831) and the corresponding region of trypanosomatid CDC5 sequences is indicated by the dashed box. PRP19: U-Box (smart00504), E = 1.06e−14; PRP19 domain (coiled-coiled [C-C] protein interaction domain, pfam08606), E = 4.87e−20; WD40 (cd00200), E = 5.77e−18. PRL1: WD40 (cd00200), E = 1.4e−75. SPF27: while the LPY motif is present in T. brucei SPF27, the human BCAS2 domain (pfam05700) was not recognized. PRP17: WD40 (cd00200), E = 2e−37. SKIP: PPIL1-binding domain (PBD); SKIP_SNW domain (pfam02731), E = 7.51e−08. PPIL1: cyclophilin domain [cd00317], E = 2.18e-27 (note that while the trypanosome sequence returned the general cyclophilin domain, the more specific SpCYP2_like cyclophilin domain (cd01922) was identified in the human ortholog.
B. Multiple sequence alignment of the SKIP/SNW domain of Homo sapiens (Hs, accession number NP_036377), Drosophila melanogaster (Dm, AGB95213), Caenorhabditis elegans (Ce, CAA98552), Arabidopsis thaliana (At, AEE35947), Schizosaccharomyces pombe (Sp, CAB41231), Saccharomyces cerevisiae (Sc, P28004), and the kinetoplastids Leishmania major (Lm, LmjF.15.1030), Leishmania braziliensis (Lbr, LbrM.15.1070), Trypanosoma cruzi (Tc, TcCLB.509445.20), T. brucei(Tb, Tb927.9.5880), Trypanosoma vivax(Tv, TvY486_0902160), and Bodo saltans (Bs, ACI16065). Since the N-terminal ~ 50 amino acids of the domain are only weakly conserved among kinetoplastids, they were omitted from the alignment. Numbers indicate the position within the SKIP/SNW domain. Positions with more than 50% similarity or identity are shaded in gray and black respectively. Identical positions in model organisms without any conservation in kinetoplastids are shaded blue and identical position in kinetoplastids without conservation in model organisms are shaded in red. A hyphen indicates lack of an amino acid at this position. Numbers in parentheses specify lengths of non-conserved insertions. The highly conserved SNWKN motif is indicated by asterisks.
C. Corresponding alignment of four short conserved domains in SPF27 orthologs. Asterisks mark the highly conserved LPY motif, which, among Leishmania species, is only present in L. tarentolae as LPF. Accession numbers: Hs, NP_005863; Dr(Danio rerio), NP_001007775; Dm, NP_651596; Ce, NP_498360; At, NP_566599; Sp, CAB57933; Sc, EEU07188; Ddis (Dictyostelium discoideum), XP_640072; Tb, Tb927.11.14150; Tcon (Trypanosoma congolense), TcIL3000.11.14450; Tv, TvY486_1114990; Tc, TcCLB.511727.110; Lta (Leishmania tarentolae), LtaP32.0970; Lm, LmjF.32.0900.
D. Multiple sequence alignment of the 21 amino acid-long PPIL1 binding domain in SKIP orthologs. Numbers indicate positions relative to the starting methionine. The two key residues for PPIL1 binding are marked by asterisks.