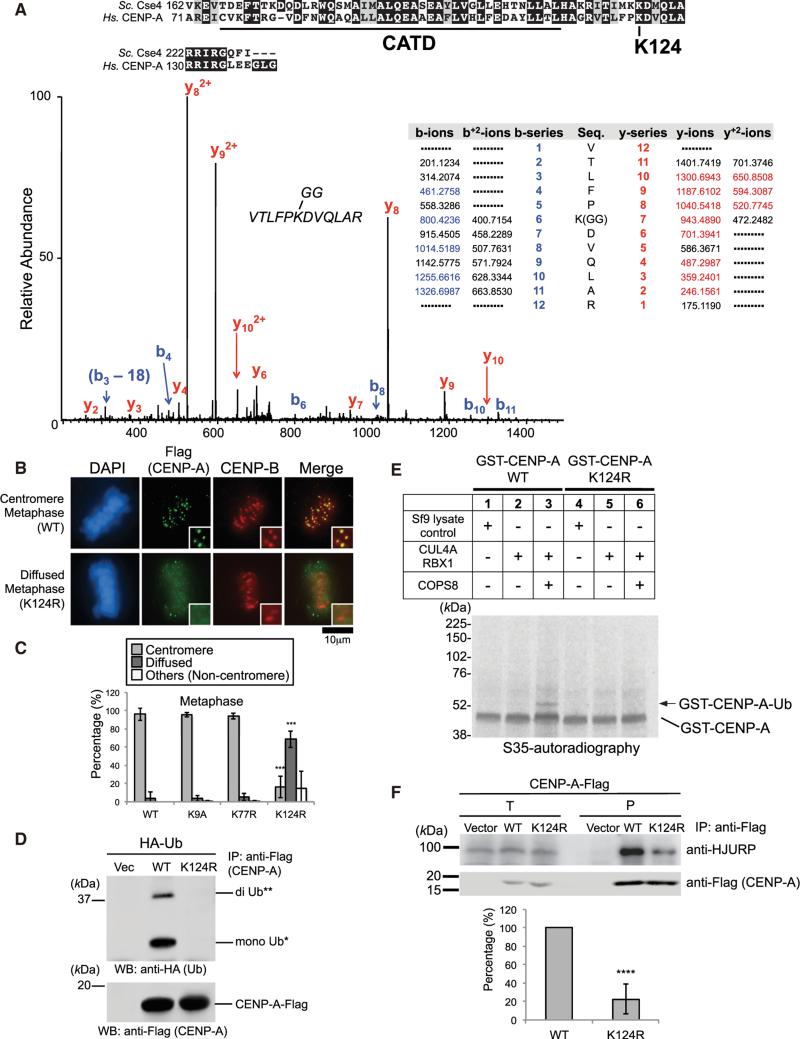
Figure 3. CENP-A K124 Ubiquitylation Is Required for CENP-A Localization at Centromeres.
(A) (Top) C-terminal conserved region between S. cerevisiae Cse4 and human CENP-A. The ubiquitylation site (lysine 124) on human CENP-A is indicated, and the CATD is underlined. (Bottom) Lysine 124 on CENP-A is a ubiquitylation site in vivo. Collision-induced dissociation analysis of the VTLFPKGGDVQLAR (m/z 500.94) peptide is displayed (coverage 55%). The b (blue) and y (red) ions detected during fragmentation are highlighted in the table. The y-7 ion (m/z 943.4890) confirms the modification of K124 by the di-glycine motif (see Table S7).
(B) The CENP-A K124R mutant delocalizes from centromeres (see Immunofluorescence in Supplemental Experimental Procedures). DAPI (blue), Flag (green), and endogenous CENP-B (red) as a centromere location control were visualized. Note that diffused signals appear in the exogenous CENP-A-Flag over-expression, presumably because its expression level is approximately 1.0–1.4 orders of magnitude (10- to 25-fold) higher than endogenous CENP-A (data not shown). Representative images of other cell-cycle stages are shown in Figure S4D. Scale bar, 10 μm.
(C) Histograms summarizing the localization patterns shown in (B). More than 50 metaphase cells were counted per experiment (n ≥ 3 experiments), and the mean percentages (±SD) are shown. “Others (Non-centromere)” indicates mostly damaged cells, dead cells, or cells with nucleolar localization (only in inter-phase) presumably due to transfection or other treatments. Histograms of other cell-cycle stages are shown in Figure S4E. ***p < 0.001 compared with CENP-A WT-Flag (Student's t test).
(D) The CENP-A K124R mutation abrogates ubiquitylation of CENP-A in vivo (see Experimental Procedures). WT, CENP-A (WT)-Flag; K124R, CENP-A (K124R)-Flag.
(E) The CENP-A K124R mutation abrogates ubiquitylation of CENP-A in vitro (see Experimental Procedures). 6xHis-tagged components (CLU4A, RBX1, and COPS8) are shown in the upper table. Band of putative GST-CENP-A-Ub is indicated with arrow.
(F) (Top) The CENP-A K124R mutation reduces the interaction with HJURP in vivo (see Immunoprecipitation Assay in Supplemental Experimental Procedures; see also Discussion and Figures S5E and S7E). Proteins in 2% of the total cell lysates (T) and immunoprecipitates (P) were detected. (Bottom) Histogram shows quantified coprecipitated endogenous HJURP bands represented in (top) (ratio to Flag band signal normalized with pTRM4-CENP-A WT-Flag-transfected cells [left column]). Experiments were repeated (n ≥ 4 experiments), and mean percentages (±SD) are shown. ****p < 0.0001 compared with pTRM4-CENP-A WT-Flag-transfected cells (left column, Student's t test).