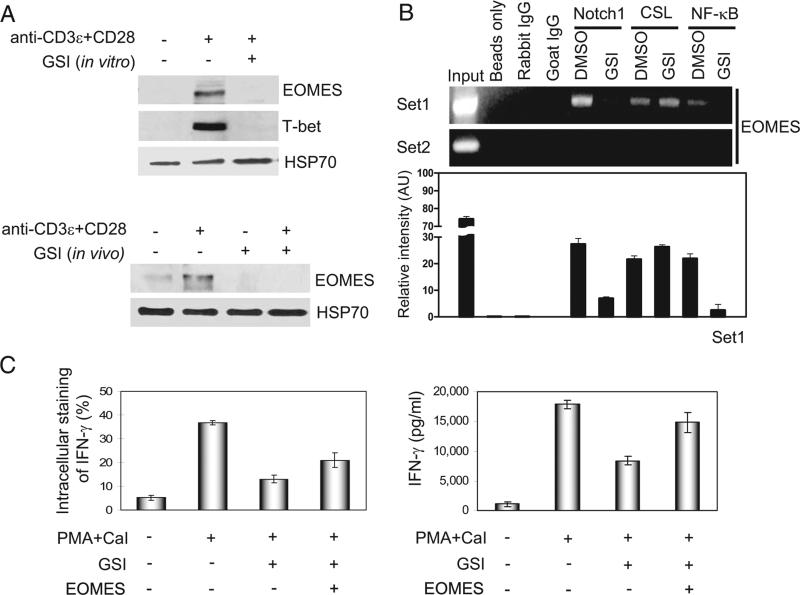
FIGURE 7.
Notch1 signaling directly regulates EOMES expression in GSI-treated CD8+ T cells. A, For in vitro treatment of CD8+ T cells (top blots), splenocytes were harvested from C57BL/6 mice and CD8+ T cells were purified before pretreating them in vitro with DMSO or GSI, stimulating them for 2 days with anti-CD3ε plus anti-CD28, and analyzing whole cell lysates by immunoblotting for EOMES and T-bet expression. For in vivo treatment of CD8+ T cells (bottom blots), control chow, or GSI formulated in rodent chow, was fed to C57BL/6 mice for 13 days. We then harvested splenocytes, purified CD8+ T cells, and stimulated these cells for 2 days with anti-CD3ε plus anti-CD28 before analyzing whole cell lysates by immunoblotting for EOMES expression. B, Two sets of PCR primers (set1 and set2) were designed to include putative binding sites for CSL and NF-κB and ChIP analysis of the EOMES promoter was performed using purified CD8+ T cells pretreated with DMSO or with GSI before stimulation for 2 days with anti-CD3ε plus anti-CD28 (top bands). Whole cell lysates were immunoprecipitated using anti-Notch1, anti-CSL, or anti-p50; rabbit and goat isotype IgG were used as negative controls. For PCR, we used 5 μl of DNA elutes. Results using primer set2 suggest that binding to putative sites within the EOMES promoter is specific. We quantified the relative intensity of the bands at the top, as represented graphically (bottom panel), which represents the mean intensity ± SD of three independent experiments. C, Exogenous EOMES was overexpressed in purified CD8+ T cells, and intracellular staining (left) and ELISA (right) were used to measure the ability of EOMES to induce IFN-γ in CD8+ T cells pretreated with GSI. PMA and calcium ionophore (CaI)-treated (PMA+CaI) and CD8+ T cells retrovirally infected with pMX-EOMES-IRES-hCD8 (EOMES +) or CD8+ T cells retrovirally transduced with empty vector of pMX-IRES-hCD8 as control (EOMES +) are also indicated. Data represent three or four independent replicates.