Abstract
Eukarya pyruvate kinases have glutamate at position 117 (numbered according to the rabbit muscle enzyme), whereas in Bacteria have either glutamate or lysine and in Archaea have other residues. Glutamate at this position makes pyruvate kinases K+-dependent, whereas lysine confers K+-independence because the positively charged residue substitutes for the monovalent cation charge. Interestingly, pyruvate kinases from two characterized Crenarchaeota exhibit K+-independent activity, despite having serine at the equivalent position. To better understand pyruvate kinase catalytic activity in the absence of K+ or an internal positive charge, the Thermofilum pendens pyruvate kinase (valine at the equivalent position) was characterized. The enzyme activity was K+-independent. The kinetic mechanism was random order with a rapid equilibrium, which is equal to the mechanism of the rabbit muscle enzyme in the presence of K+ or the mutant E117K in the absence of K+. Thus, the substrate binding order of the T. pendens enzyme was independent despite lacking an internal positive charge. Thermal stability studies of this enzyme showed two calorimetric transitions, one attributable to the A and C domains (Tm of 99.2°C), and the other (Tm of 105.2°C) associated with the B domain. In contrast, the rabbit muscle enzyme exhibits a single calorimetric transition (Tm of 65.2°C). The calorimetric and kinetic data indicate that the B domain of this hyperthermophilic enzyme is more stable than the rest of the protein with a conformation that induces the catalytic readiness of the enzyme. B domain interactions of pyruvate kinases that have been determined in Pyrobaculum aerophilum and modeled in T. pendens were compared with those of the rabbit muscle enzyme. The results show that intra- and interdomain interactions of the Crenarchaeota enzymes may account for their higher B domain stability. Thus the structural arrangement of the T. pendens pyruvate kinase could allow charge-independent catalysis.
Introduction
Rabbit muscle pyruvate kinase (RMPK) was the first enzyme reported to have an absolute requirement for K+ [1]. Despite extensive study [2,3,4,5,6], the role of the K+ in catalysis by RMPK is not yet completely understood. Recently, it has been proposed that the K+ is directly involved in the movement of the active site lid (B domain) during the transition of PK to its active conformation. This conformation allows either phosphoenolpyruvate (PEP) or ADP to bind, following a random-order kinetic mechanism [7]. For a long time, it was thought that the dependence on K+ was a feature common to all PKs [8]. However, as more enzymes were characterized, it became apparent that the activity of many PKs is K+ independent [8]. To explore the molecular basis underlying this behavior, Laughlin and Reed [9] compared the amino acid sequence of RMPK with those of two K+-independent bacterial enzymes. These authors found that Glu117 of RMPK, which is close to the K+-binding site, was replaced by Lys in the bacterial enzymes. They constructed the E117K mutant of the rabbit enzyme and found that the mutant was not stimulated by monovalent cations. The authors proposed that the expression of the K+-independent activity was due to the internal positive charge supplied by the protonated Lys [9]. To determine the abundance of K+-independent PK enzymes, an extensive phylogenetic study of this enzyme family was performed [8]. Of the 230 sequences investigated, 121 contain a Glu at position 117 (according to RMPK numbering), 106 contain a Lys, 2 contain a Ser and 1 has an Arg at this position. The PKs containing a Glu and those containing a Lys are clearly separated into two clusters. All characterized members of the cluster with a Glu residue at position 117 exhibit K+-dependent activity, whereas those with a Lys residue exhibit K+-independent activity. The presence of Leu113/Gln114 and a hydrophobic residue (Ile, Leu, Val) at position 120 are covariant in 77% of the PKs that contain Lys117. These residues are replaced by Glu117/Thr113/Lys114/Thr120 in 80% of the K+-dependent PKs.
Laughlin and Reed [9] also constructed an E117A mutant of RMPK and found that, like the wild-type, its activity was absolutely dependent on K+. This result confirmed their hypothesis that the enzyme requires an internal positive charge or K+ for catalytic activity. However, the PKs from Pyrobaculum aerophilum (PaPK) and Thermoproteus tenax are K+-independent even if a non-positively charged residue (Ser) is located at the position that corresponds to residue 117 of RMPK [10,11]. These two homologs are located in the K+-independent branch of the phylogenetic tree of PKs [8] and belong to the Crenarchaeota subdomain. The phylogenetic analysis previously described, included only 18 sequences of the PK proteins from Archaea, and the number of sequences of PKs available is substantially larger today. In contrast to the conserved Glu117/Lys117 present in all PKs from the Bacteria and Eukarya domains, other amino acids are found in the corresponding position in the enzymes from the Crenarchaeota subdomain. In this work, an updated sequence alignment of the Archaeal PKs is presented (S1 Fig.). The previously observed co-evolution between residue 117 and residues 113, 114, and 120 [8] was not observed in the sequences of PKs from hyperthermophilic Crenarchaeota species, nor were the residues of the K+ binding site strictly conserved.
These data suggest that the K+-independent activity in Crenarchaeota PKs may result from a mechanism other than that involving the internal positive charge supplied by the protonated ε-amino group of Lys [9]. To further investigate these hyperthermophilic enzymes, we performed a new phylogenetic analysis, evaluated the kinetic properties, thermal stability, and molecular dynamics, and performed structural modeling of the pyruvate kinase from Thermofilum pendens (TpPK), which has a hydrophobic residue at the position corresponding to the RMPK 117 (Val70). The results indicate that for TpPK, the closure of the active site and the arrangement of the residues involved in the binding of the nucleotide are independent of the presence of an internal positive charge or K+ and may be related to the stability of the active site cleft.
Materials and Methods
Reagents
The endonucleases and T4 DNA ligase were purchased from New England Biolabs (UK). Platinum Taq DNA polymerase was from Invitrogen Life Technologies (MA, USA). Shrimp alkaline phosphatase was from Roche Applied Science. Miniprep plasmid purification kits were from QIAGEN (N.V.). The BL21 CodonPlus-pLysS strain of E. coli, the pMCSG7, pET-3a vectors and all oligonucleotides were from Invitrogen. RMPK and hog muscle lactate dehydrogenase (LDH) were obtained from Roche Applied Science (Mannheim, Germany). Antibiotics (ampicillin, chloramphenicol), Mes, Tris and the cyclohexylammonium salts of ADP and PEP (phosphoenolpyruvate) were from Sigma-Aldrich Co. (USA) NADH sodium salt was converted to the cyclohexylammonium salt by ion exchange following the protocol provided by the manufacturer (Sigma-Aldrich).
Sequence analyses
The pyruvate kinase amino acid sequences from Archaea were retrieved by BlastP searches at the UniProt site [12] (http://www.uniprot.org). Progressive multiple amino acid sequence alignments were performed with ClustalX version 2 [13] (http://www.clustal.org/clustal2/) using a structural alignment constructed with the VAST algorithm as a guide [14]. The alignment included all non-redundant PK protein structures deposited in the Protein Data Bank [15]. Non-redundant protein sequences from a previous phylogenetic analysis [8] were included in the multiple-sequence alignment and corrected manually using BioEdit [16] (http://www.mbio.ncsu.edu/bioedit/bioedit.html). Phylogenetic analyses were conducted using the MEGA5 software [17] (http://www.megasoftware.net). Four methods were used to infer the phylogenetic relationships: maximum likelihood, maximum parsimony, minimum evolution, and neighbor joining. The amino acid substitution model described by Whelan and Goldman [18] using a discrete Gamma distribution with five categories was chosen as the best substitution model because it gave the lowest Bayesian Information Criterion values and corrected Akaike Information Criterion values [19] in MEGA5 [17]. The gamma shape parameter value (+G parameter = 0.92) was estimated directly from the data with MEGA5. The confidence for the internal branches of the phylogenetic tree, obtained using the maximum likelihood method, was determined through bootstrap analysis (500 replicates each).
Sequence logos were constructed using the WebLogo server (http://weblogo.threeplusone.com/). Each logo consists of stacks of amino acid letters. The ordinate axis of the logos graphs, indicate the stack for each position in the sequence. The height of the letters within the stack indicates the relative frequency of each amino acid at that position [20].
Cloning of TpPK
The PK gene from T. pendens was amplified from the genomic DNA of the strain Hrk. The gene was amplified by PCR using the primers FW 5′-TACTTCCAATCC AATGCTGCAAAAGTCAAGCTAGTAGCGCG-3′ and RV 5′-TTATCCACTTCCAATGTT ACTCGCTCTTATCTCTCCACGG-3′. The insert was introduced into the pMCSG7 vector, a pET-based expression vector that has a His6 tag at the N-terminus and a Tobacco Etch Virus (TEV) protease cleavage site, by ligation-independent cloning using published protocols [21]. The vector was treated with SspI followed by T4 DNA polymerase in the presence of dGTP, and the PCR product was treated with polymerase in the presence of dCTP. After annealing, freshly prepared competent Escherichia coli XL-Gold were transformed with the plasmids. The plasmids were isolated and sequenced to verify the absence of mutations.
Cloning of the B domain
The fragment encoding the B domain was amplified from the TpPK plasmid by PCR, using the following primers FW 5′-TACCATATGAGGCTTGGAGAG-3′ and the RV 5′-ATTGGATCCGACGGTCACAGT-3′. The PCR product was cloned into the pET-3a vector using two restriction sites (Nde1 and BamH1). The pET-3a vector was transformed into E. coli XL-Gold. The plasmids were isolated and sequenced to verify the absence of mutations.
Cell growth and purification of TpPK
LB medium containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol was inoculated with BL21 cells containing the plasmid CodonPlus-pLysS-TpPK at 37°C at an A 600 of approximately 0.4. Expression was induced overnight with 0.5 mM isopropyl 1-thio-β-D-galactopyranoside at 15°C. Recombinant E. coli cells were suspended in 50 mM KH2PO4 pH 8, 10 mM Imidazol, 300 mM KCl and half of a tablet of Complete Protease Inhibitors (Roche Applied Science). The cells were lysed by sonication with a Sonifier 450 (Branson) for 2.5 min at 40 kHz. The suspension was centrifuged, the supernatant was heated at 80°C for 30 min, and the precipitated protein was discarded. The supernatant was loaded on a His Trap FF column, and the enzyme was eluted with a linear gradient of imidazole (10–500 mM). The fractions that exhibited PK activity (~ 250 mM imidazole) were pooled and concentrated by membrane filtration (Centricon 30,000 MWT) and then desalted on a Hi Trap Desalting column. This pool was incubated for 48 h with Tobacco Etch Virus (TEV) protease at a ratio 1:30 (TpPK: TEV). The enzyme was then loaded on a His Trap column. The fractions with activity were pooled and concentrated. In this step, less than 0.5% of the TpPK was recovered. Therefore, in subsequent purifications, the incubation with TEV protease was omitted. The enzyme that was still bound to the His Trap FF column was eluted with a linear gradient of imidazole. The fractions that exhibited PK activity were pooled, concentrated (Centricon 30,000 MWT) and desalted on a Hi Trap Desalting column. This pool was loaded on a DEAE Sepharose column, and the enzyme was eluted with a linear gradient of KCl (0–1 M). The purity of the TpPK that eluted from the imidazole gradient was 89% (in this step, the purity of TpPK with or without His6 tag was similar; gel not shown), whereas that of the enzyme after the anionic-exchange step was >95% as determined by SDS-PAGE (12.5%). Mass spectrometric characterization of the protein by ESI-MS was performed at the Research Resources Center at the University of Illinois, Chicago (spectrum not shown). The enzyme was precipitated with ammonium sulfate at 80% saturation and stored at 4°C. To determine the oligomerization state of the enzyme, BN-PAGE was carried out overnight at 4°C as described [22] using 150 μg of TpPK.
Cell growth and purification of the B domain
LB medium containing 100 μg/ml ampicillin and 34 μg/ml chloramphenicol was inoculated with BL21 cells containing the plasmid CodonPlus-pLysS-B domain at 37°C at an A 600 of approximately 0.4. Expression was induced with 0.5 mM isopropyl 1-thio-β-D-galactopyranoside at 20°C, and the cells were harvested after overnight growth. Recombinant E. coli cells were suspended in 50 mM Tris-HCl pH 7.0 containing half of a tablet of Complete Protease Inhibitors (Roche Applied Science). The cells were lysed by sonication with a Sonifier 450 (Branson) for 2.5 min at 40 kHz. The suspension was centrifuged 30 min at 20,000 X g. The supernatant was precipitated with ammonium sulfate at 37% saturation, and the resultant supernatant was collected. A second precipitation with ammonium sulfate at 80% saturation was performed. The pellet was collected, suspended and desalted by dialysis. The final steps involved ion exchange chromatography using DEAE and molecular exclusion using Superdex 75. SDS-PAGE was performed, and the fractions with MW ~10,000 Da were pooled and concentrated. The B domain was 95% pure, as indicated by SDS-PAGE (not shown).
Mass spectroscopy of the B domain of TpPK
The protein band with the expected molecular weight of the B domain was excised from the SDS-PAGE gel. The sample was digested in-gel with trypsin and was then injected into an integrated nano-LC-ESI-MS/MS system (quadrupole/time of flight, Ultima API, Micromass, Manchester, UK). The acquired peptide ions were analyzed with the Mascot program (www.matrixscience.com) using both the NCBInr and the EST databases. Only proteins with significant ion scores (> 46) were reported. The sequence of all peptides matched the B domain of TpPK (data not shown). These experiments were performed by Dr. Maire Christine Slommiary of Université des Sciences at Technologies de Lille, Villeneuve d′ Ascq, France.
Assays of pyruvate kinase activity
LDH was obtained as an ammonium sulfate suspension from Roche Applied Science. Ammonium sulfate-free enzymes were obtained as described previously [23]. Contaminating NH4 +, Na+, and K+ in the reaction mixtures were below the detection limit (10 μM), as indicated elsewhere [24]. The formation of pyruvate was measured at 45°C in a coupled system with LDH and NADH [25]. The reaction mixtures contained 50 mM Mes-Tris, pH 6.0 and the indicated concentrations of cations (Mg2+ and Mn2+), substrates (PEP and ADP), and inhibitors (oxalate and AMP). The ADP-Mg complexes and free Mg2+ concentrations were calculated using the software CHELATOR [26]. The ADP-Mn complexes and free Mn2+ concentrations were calculated using the K d of Mn2+ [27]. The ionized PEP concentrations were calculated considering a pK value of 6.3 [28]. (CH3)4NCl was added to a final salt concentration of 0.25 M to compensate for the different ligand (PEP, ADP-Mg, ADP-Mn, free divalent cations) concentrations. In the inhibition assays, the concentration of LDH added to the reaction mixture was sufficient to overcome the inhibition by oxalate. The specific activity was not increased by the inclusion of 5-fold higher concentrations of LDH. The reaction mixture was incubated for 10 min to reach the desired temperature (45°C) prior to initiating the reaction with TpPK.
Kinetic studies
The initial velocities of TpPK activity were determined in the absence or presence of dead-end inhibitors (oxalate or AMP). In the former condition, the velocity patterns were obtained at various concentrations of PEP at several fixed concentrations of ADP-Mg. In the latter condition, the inhibition patterns were obtained by varying the concentration of one substrate with the second substrate fixed and at different fixed concentrations of the inhibitor.
Differential scanning calorimetry (DSC)
All experiments were performed in a capillary Differential Scanning Microcalorimeter from GE Health Science (USA). The protein solutions were prepared in 50 mM Tris-HCl pH 7.6. The buffer in which the enzyme was prepared was placed in the reference cell. Unless otherwise indicated, the protein concentration was 1 mg/ml and the scan rate was 1.5°C/min. The experiments were conducted at temperatures ranging from 25 to 120°C.
Models and structural analysis
The structures for RMPK (PDBID 2G50 was used as the open form, and 1A5U was used as the closed form) and PaPK (PDBID 3QTG) were obtained from the PDB database (RCSB). The structures of TpPK and the mutant F89I/F108I/F109C/F127L of TpPK were modeled with Modeller version 9.2 [29] using the structure of PaPK (the only hyperthermophilic PK structure from Crenarchaeota phylum that has been determined) as the template [30]: the identity between the two sequences was 30.6%. The model of the structure of TpPK was validated with Molprobity (S1 Table) [31]. H-bonds were quantified using the Find H-Bond module in UCSF Chimera version 1.8.1 [32], and salt bridges were counted by first selecting the atoms involved in this type of interaction (Ng+, N3+, N2+, O2-, O3-) and then counting the interactions using the same Find H-bond module. π-π and π-cation interactions were counted using Yasara version 18.9.8 (“Yet Another Scientific Artificial Reality Application,” http://www.yasara.com) with the view interactions module. Specific interactions between domain A and domain B were identified by using Dimplot 4.5.3 [33] and its domain-domain interface module with the following definitions for each domain: domain B 66–164 (TpPK), 81–177 (PaPK) and 116–223 (RMPK); domain A 1–65 and 165–334 (TpPK), 1–80 and 178–346 (PaPK) and 43–115 and 224–387 (RMPK). Molecular dynamic simulations were run using AMBER12 [34]. The models were prepared with tleap using the AMBER FF12SB force-field and an implicit solvent model. Briefly, the system was first energy-minimized with 5000 steps of steepest descent followed by 5000 steps of conjugate gradients. Then, the system temperature was raised from 0 to the final simulation temperature (300, 400 or 500 K) over 200 ps. The production simulations (50 ns) were started after this step. For all simulation steps, a Langevin thermostat was used with a collision frequency of 1 per picosecond, and SHAKE was applied only to the hydrogen atoms with a step time of 2 femtoseconds. All simulations were run on GPUs [35]. Analysis of the native contacts was calculated using Carma [36,37].
Results and Discussion
Phylogenetic analysis of PKs
An updated bioinformatic analysis of PK [8] that included 191 new protein sequences from Archaea was performed. The new analysis consisted of 426 non-redundant PK sequences: 71 from Eukarya, 151 from Bacteria and 204 from Archaea. It is noteworthy that, without exception, the PKs from Archaea corresponded to single-copy genes. S1 Fig. shows that the PK protein sequences are divided into two main clusters: those with Glu at position 117 (according to RMPK numbering), all of which are K+- dependent, and those that mainly contain Lys at the corresponding position, all of which are K+- independent [8]. The analysis of the logos shows that position 117 covaries with positions 113, 114 and 120 (S1 Fig.).
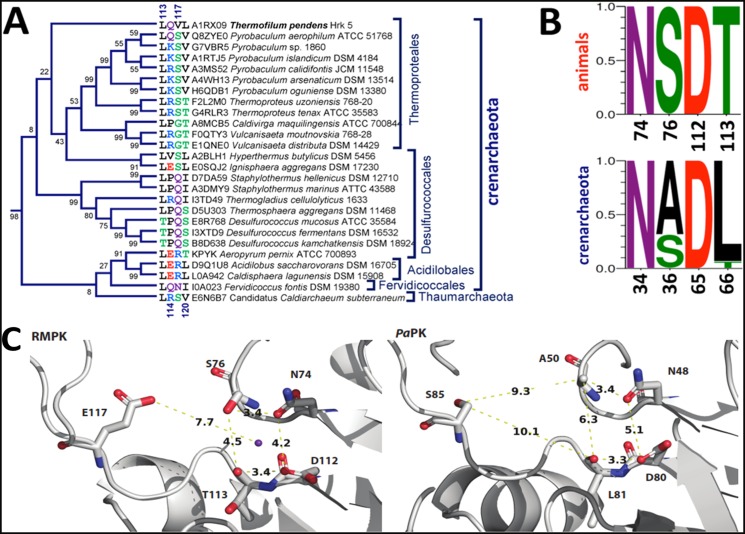
The TpPK sequence is clustered with the other Crenarchaeota PKs in a branch that includes PK sequences from thermoproteales, desulfurococcales, acidilobales and fervidicoccales (Fig. 1A). This branch is located within the cluster that comprises the K+-independent enzymes (see S1 Fig.). However, this branch contains the sequences of PKs in which residues other than Lys at position 117 are present (Ser, Gln, Arg, Asn, Gly and Val) (Fig. 1A and S1 Fig.). For instance, TpPK has Val70 at the equivalent position, whereas the PKs from other Crenarchaeota species, such as Pyrobaculum aerophilum and Thermoproteus tenax, have Ser at this position and are also K+-independent [10,11]. This finding suggests that catalysis by these enzymes utilizes a mechanism other than that of an internal positive charge provided by Lys. Interestingly, in this Crenarchaeota branch, the covariation of the residues at positions 114, 117 and 120 is not conserved. In contrast, PaPK and TpPK are the only two Crenarchaeota PKs that share the covariation of residues at positions 113, 114 and 120 with the K+-independent enzymes. The other sequences of the PKs in this unusual Crenarchaeota branch share only one or two residues of the K+-independent signature. As described in [8], the residues that form the K+ binding site are highly conserved either in the K+-dependent or-independent PK branches, with the exception of Thr113, which is almost exclusively observed in the K+-dependent PKs. In the Crenarchaeota branch, only two of the four oxygens that putatively coordinate with the K+ are conserved (Oδ1 of Asn34 and Oδ2 of Asp65), and only 30% of the sequences contain the third one (Oγ of Ser36) (Fig. 1B). This result indicates that these residues are not sufficient to coordinate the K+. A comparison of the K+-binding site in RMPK with the putative monovalent site in PaPK is shown in Fig. 1C. Remarkably, the distances between the oxygen atoms of the residues involved in the coordination sphere of K+ in RMPK are shorter than those found in the corresponding residues in PaPK. Moreover, the distance between O∂2 of Glu117 and Oγ of Ser76 is 5.1 Å in RMPK, whereas the distance between Oγ of Ser85 and Cß of Ala50 is 9.3 Å in PaPK. This observation indicates that Glu117 is close to the K+-binding site and is a key residue in the dependence of RMPK on monovalent cations. In contrast, in PaPK, this position (Ser85) is probably too far away to affect this site. It is relevant to consider that the rotation angle of the B domain relative to the A domain is 41° for RMPK (PDBID 1A49, B subunit). This subunit exhibits the most open cleft conformation reported for RMPK [38]; the distance from O∂2 of Glu117 to Oγ of Ser76 is 5.1 Å, and the distance to the carbonyl oxygen of Thr113 is 7.7 Å (not shown). These distances are shorter than the distances shown for PaPK in Fig. 1C. Although a strict comparison between these two crystal structures (RMPK vs. PaPK) cannot be made due to the lack of information concerning the rotation angle of the B domain over the A domain for PaPK (PDBID 3QTG) [30], Ser cannot occupy the same site that Glu does due to the differences in the sizes of the two residues.
Fig 1. Conservation of the K+ binding site residues and of those that covariate with the corresponding position 117 in Crenarchaeota subdomain.
In A. the subtree of the Crenarchaeota branch that contains the TpPK sequence is shown. The unrooted phylogenetic tree that includes all available PK protein sequences from the Archaea domain is shown in Supplemental data (S1 Fig.): here, only the subtree of the Crenarchaeota branch that included TpPK is presented. Residues 113, 114, 117, and 120 (according to RMPK numbering), as well as the accession numbers are indicated. The branches are not drawn to scale, and only the branch topology is shown. B. Logos of the K+ binding site residues in PK from animals and from the Crenarchaeota subdomain are presented. In animals, the numbering is according to RMPK, and the equivalent positions in the Crenarchaeota subdomain are given according to TpPK numbering. C. The K+ binding site in RMPK (PDBID 1A49, subunit B) and hypothetical residues of coordination to the monovalent cation in PaPK (PDBID 3QTG). In RMPK, K+ is shown in purple and the distances from K+ to Oγ of Ser76, O∂1 of Asn74, O∂1 of Asp112 and the carbonyl oxygen of Thr113 are 3.1, 2.6, 2.6 and 2.8 Å, respectively (not shown). The distances shown are those between the coordination residues and from O∂2 of Glu117 to K+ (RMPK) or from Oγ of Ser85 to Cß of Ala50 and the carbonyl oxygen of Leu81 (PaPK).
Purification of TpPK and molecular mass determination
TpPK was purified as described previously (Material and Methods) and incubated in the presence of TEV protease; however, the recovery of the total TpPK without the His6 was less than 0.5%. Therefore, unless otherwise indicated, the experiments were performed with the enzyme containing the 23 additional residues. In this preparation, a single Coomassie-stained band with an apparent molecular mass of 50 kDa was observed by SDS-PAGE, confirming its high degree of purity (S2A Fig.). The oligomeric state of TpPK was determined by BN-PAGE [22], which showed a native band of approximately 200 kDa (S2B Fig.), indicating that like most of the known PKs, the TpPK is a homotetramer [39]. Mass spectrometry of the enzyme yielded a molecular weight of 53,965 Da, with 51,340 Da corresponding to the monomer of TpPK and 2,624.7 Da corresponding to the His6 tag and the cleavage site of protease TEV (data not shown).
Catalytic Properties of TpPK
Because there was no previous information available about TpPK, the enzyme was characterized biochemically. The kinetic constants for ADP-Mg of TpPK with and without His6 tag were similar (Km of 0.092 ± 0.012 mM and 0.14 ± 0.013 mM, and V max of 175 ± 6 μmol/min mg and 176 ± 5 μmol/min mg without and with the His6 tag, respectively), ruling out any putative effect of the 23 extra residues on the kinetic behavior of TpPK.
Effect of monovalent cations
To date, three PKs from Crenarchaeota species (Pyrobaculum aerophilum, Thermoproteus tenax and Aeropyrum pernix) have been characterized [10,11]. The first two contain Ser85 and Ser69, respectively (corresponding to position 117 in RMPK), the last contains Arg72, and these enzymes express monovalent cation-independent activity. Consistent with the PKs from Pyrobaculum aerophilum and Thermoproteus tenax, TpPK that has a non- positively charged residue at the position corresponding to residue 117 and exhibited K+-independent activity as indicated by absence of an increase in activity when monovalent cations were included in the reaction mixture (data not shown). The lack of activation by monovalent cations has been reported in all K+-independent enzymes, including those with Lys [40,41,42,43,44,45,46], Ser [10,11] or Arg [11] at the position corresponding to residue 117.
Kinetics of TpPK in the presence of Mg2+ and Mn2+
The effects of divalent metal ions on the catalytic activity of TpPK were studied because divalent cations are essential for phosphate transfer [47]. Therefore, in the absence of monovalent cations, we explored the effect of the “ancient Mn2+” [48] and of Mg2+, the physiologically relevant divalent metal ion. The kinetic constants in the presence of these two divalent cations are shown in Table 1. Although the maximum activity was 3.8-fold higher with Mg2+ than with Mn2+, the K 0.5 for Mn2+ was 250-fold lower than the K 0.5 for Mg2+, with no significant change in the constants for the substrates. This finding shows that Mn2+ is the preferred divalent cation, consistent with the geochemistry of the Archaean ocean [48]. The K 0.5 for Mn2+ of TpPK (20 μM) is one of the smallest reported to date: the K 0.5 for Mn2+ of the PK of Thermoproteus tenax is 800 μM, whereas the K 0.5 for Mg2+ was the same for both enzymes (~5 mM) [10]. In addition, the PK of Thermoproteus tenax exhibited the same V max (45 μmol min-1 mg-1) at 50°C in the presence of either Mn2+ or Mg2+ [10].
Table 1. Kinetic constants for TpPK in the presence of Mg2+ and Mn2+.
| Mg2+ | Mn2+ | |||
|---|---|---|---|---|
| K m | n | K 0.5 | n | |
| PEP 3- (mM) | 0.38 ± 0.03 | ___ | 1.0 ± 0.1 | 1.2 ± 0.1 |
| ADP- M 2+ (mM) | 0.16 ± 0.01 | ___ | 0.2 ± 0.02 | 1.2 ± 0.3 |
| M 2+ (mM) | 5.0 ± 0.28 | 1.8 ± 0.4 | 0.02 ± 0.001 | 1.6 ± 0.2 |
| V max (μmol/ min • mg) | 183 ± 8 | 41 ± 1.9 | ||
The data with Mg2+ from Fig. 2 were globally fit (nonlinear regression, Origin version 6.0) to the equation describing a rapid-equilibrium random-order mechanism: v = V max[A][B]/(K a K b + K a [B] + K b[A] + [A][B]), where v represents the initial velocity, A is PEP, B is ADP-Mg, and K a and K b are the Michaelis-Menten constants for PEP3- and ADP-Mg, respectively. S.D. values are shown. The data (not shown) with Mn2+ were fitted to the Hill equation v = V max*[S]n/K 0.5 n+[S]n, where S indicates the substrate concentration of PEP3-, ADP-Mn or Mn2+. The mean and standard deviations of five experiments are shown.
Effect of allosteric modulators
As for other K+-independent PKs, the binding of PEP and divalent cations to TpPK was cooperative [10,11,40,41,42]. Therefore, we explored the effects of classic allosteric modulators, such as ribose-5-phosphate, AMP and fructose-1,6-bisphosphate on the binding of PEP on the response of this enzyme. In the presence of 2 mM ADP-Mn2+ and 0.2 mM Mn2+ free, the addition of 0.5 mM ribose-5-phosphate increased the V max by 41% with minor effects on the K 0.5 for PEP and on the Hill number (S3A Fig. and S2 Table). In the presence of Mn2+, 10 mM fructose-1,6-bisphosphate increased the K 0.5 for PEP 4-fold and increased the V max by 34%, but it had no effect on the Hill number (S3B Fig. and S3 Table). In contrast, Mg2+ had no effect (not shown). AMP behaved as a competitive inhibitor of ADP-Mg with a K i of 35 ± 1.2 mM, as described in the section on dead-end inhibition studies. In contrast with other PKs from hyperthermophilic archaea, which showed no response to classic allosteric effectors [10,11], TpPK does exhibit discrete regulatory properties.
Initial velocity of TpPK
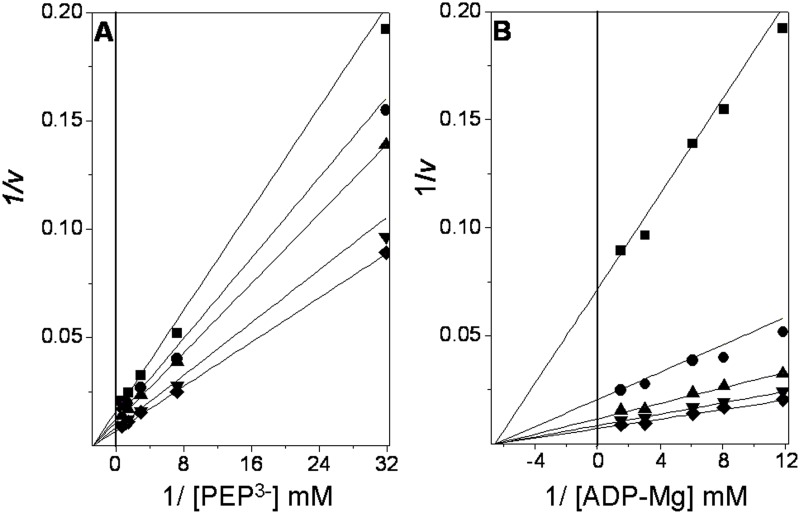
At a saturating concentration of K+, RMPK follows a random-order rapid equilibrium kinetic mechanism [49,50,51] that changes to an ordered mechanism with PEP as the first substrate in the absence of K+ [7]. In the absence of K+, the mutant E117K exhibits the same random-order rapid equilibrium kinetic mechanism [7]. These results indicate that in the muscle enzyme, the inner positive charge or K+ induces the closure of the active site and the arrangement of the residues involved in the binding of the nucleotide, allowing the random binding of PEP and ADP-Mg to the active site [7]. In this context, we were interested in determining the kinetic mechanism of TpPK. The initial velocities of the reaction of TpPK were measured without monovalent cations and with Mg2+ as the divalent cation. These experiments were carried out at various concentrations of one of the substrates and at fixed concentrations of the other (S4 Fig.). The double reciprocal plots of the initial velocities versus the ionized PEP concentrations intersected on the 1/S axis and to the left of the 1/v axis (Fig. 2A). When the concentration of ADP-Mg was varied, the lines intersected on the 1/S axis and to the left of the 1/v axis (Fig. 2B). This result indicates an ordered steady state or a random-order rapid equilibrium kinetic mechanism. The data were fit to the equation described in Table 1, and the kinetic constants obtained are summarized in Table 2.
Fig 2. Double reciprocal plots from the initial velocity data of the TpPK reaction.
The reaction medium consisted of 3 ml of 50 mM Tris-HCl pH 6.0 containing 0.24 mM NADH, 30 mM Mg2+ free, and 8 μg/ml LDH. The reciprocals of the concentrations of ionized PEP and ADP-Mg complexes are shown in the abscissas of each graph. A. The fixed concentrations of ADP-Mg were 0.084 (■), 0.12 (●), 0.16 (▲), 0.37 (▼), and 0.67 mM (♦). B. The fixed concentrations of PEP3- were 0.031 (■), 0.14 (●), 0.34 (▲), 0.69(▼), and 1.30 mM (♦). The Mg2+ free concentration was kept constant at 30 mM. To maintain the ionic strength, (CH3)4NCl was added to a final salt concentration of 0.25 M. The reaction was started with the addition of PK. The concentrations of PK were 0.32 and 0.16 μg/ml for the three lowest and two highest substrate concentrations, respectively.
Table 2. Intersecting patterns, kinetic mechanisms, and kinetic constants for TpPK.
| Initial velocity intersecting patterns | Kinetic mechanism | V max | K m PEP3- | K m ADP-Mg | k cat | Log kcat/ Km PEP3- | Log kcat/ Km ADP-Mg | |
|---|---|---|---|---|---|---|---|---|
| 1/v vs.1/PEP3- | 1/v vs.1/ADP-Mg | μmol/min·mg | mM | mM | s-1 | M-1 s-1 | M-1 s-1 | |
| Intersects to the left of the 1/v axis and on the 1/S axis | Intersects to the left of the 1/v axis and on the 1/S axis | Random rapid Equilibrium | 183 ± 8 | 0.38 ±0.03 | 0.16 ±0.01 | 626 | 6.22 | 6.59 |
Intersecting patterns were taken from the double reciprocal plots of the initial velocity data. The data shown in Fig. 2 were globally fit to the equation describing a rapid-equilibrium random-order mechanism as described in Table 1. The specificity coefficients k cat/K m (M-1 s-1) are expressed in log form.
Dead-end inhibition studies
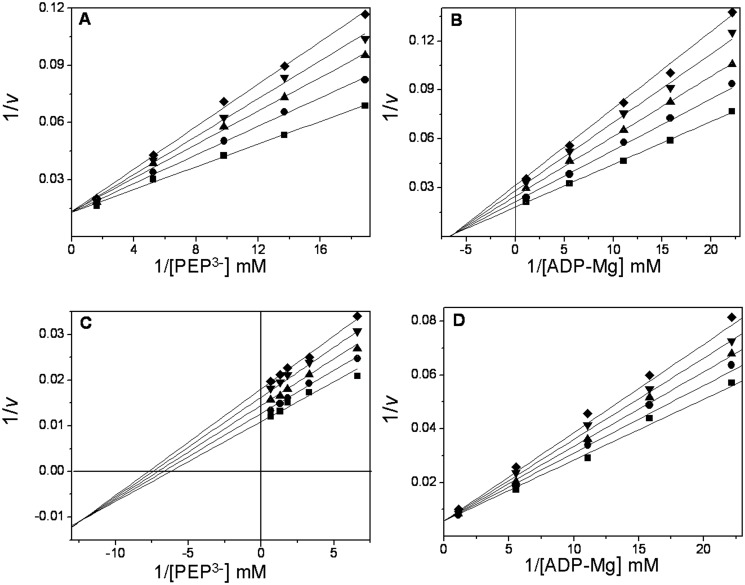
Dead-end inhibitors are powerful tools for investigating the kinetic mechanisms of enzymes [52]. Here, oxalate and AMP were used as dead-end analogs of PEP [53] and of ADP, respectively. The patterns of oxalate inhibition versus ionized PEP and ADP-Mg were competitive (Fig. 3A) and noncompetitive (Fig. 3B), respectively. With AMP, the inhibition was mixed (Fig. 3C) and competitive with ionized PEP and ADP-Mg, respectively (Fig. 3D). The data were globally fit to the equations that describe linear competitive inhibition, linear non-competitive inhibition or linear mixed inhibition. The inhibition patterns and inhibition constants are shown in Table 3. Collectively, the data show that oxalate is a competitive inhibitor with respect to PEP, whereas AMP is a competitive inhibitor with respect to ADP-Mg. This indicates that the analogs and the substrates bind to the same site. Because oxalate is a non-competitive inhibitor with respect to ADP-Mg, it may be concluded that oxalate forms a non-productive ternary complex and thereby diminishes the V max, without altering the binding of ADP-Mg. The same argument holds for AMP with respect to PEP, except that the inhibition pattern observed (Fig. 3C) was a mixed-type pattern with α < 1 (factor affecting K i) for AMP inhibition versus PEP. This difference indicated that the enzyme-AMP binary complex has a higher affinity for PEP than the free enzyme. Therefore, the results obtained using dead-end inhibitors indicate that TpPK follows a rapid-equilibrium random-order kinetic mechanism, as reported for RMPK in the presence of K+ [7,52,53,54] or the mutant E117K-PK in the absence of K+ [7]. This finding indicates that PEP and ADP-Mg bind independently to TpPK despite the absence of an internal positive charge. Similarly, it has been shown that in the presence of 40% DMSO, RMPK follows a rapid-equilibrium random-order mechanism in the absence of K+ [7]. The activity of the enzyme under these conditions is 1000-fold higher than in aqueous medium without K+ [55]. Kinetic and spectroscopic studies have shown that in the absence of K+, 40% DMSO induces the acquisition of the active (closed lid) conformation of the enzyme [55,56]. Consequently, the positive internal charge provided by Lys cannot explain the K+-independent activity observed in RMPK with 40% DMSO or in TpPK in fully aqueous media. Therefore, in accordance with the data for RMPK in DMSO and in the absence of K+, the data suggest that the overall conformation of TpPK contributes to the catalytic activity.
Fig 3. Dead-end inhibition patterns and inhibition constants for oxalate and AMP in TpPK.
The experimental conditions were as indicated in Fig. 2. The reciprocals of the concentrations of ionized PEP and ADP-Mg complexes are shown in the abscissas of each graph. In plot A, the concentrations of PEP3- were 0.054, 0.077, 0.10, 0.22, and 1.1 mM. The Mg2+ free and ADP concentrations were kept constant at 30 mM and 2 mM, respectively. The fixed concentrations of oxalate were 0 (∎), 10 (●), 20 (▴), 30 (▾) and 40 (◆) μM. In plot B, the concentrations of the ADP-Mg complexes were 0.045, 0.063, 0.090, 0.18, and 0.90 mM. The ionized PEP concentration was kept constant at 30 mM. The Mg2+ free and oxalate concentrations were as in plot A. In plot C, the concentrations of PEP3- were 0.15, 0.30, 0.55, 0.77, and 1.5 mM. The Mg2+ free and ADP concentrations were as in plot A. The fixed concentrations of AMP were 0 (∎), 4 (●), 8 (▴), 12 (▾) and (◆) 16 mM. In plot D, the concentrations of ADP were as in plot B. The Mg2+ free and ionized PEP concentrations were kept constant at 30 mM. The concentrations of AMP were 0 (∎), 8 (●), 12 (▴), 16(▾) and 20 (◆) mM.
Table 3. Dead-end inhibition patterns and inhibition constants for oxalate and AMP in TpPK.
| Dead-end analog of PEP3-: oxalate | Dead-end analog of ADP-Mg: AMP | K i (oxalate) mM | K i (AMP) mM | ||
|---|---|---|---|---|---|
| 1/v vs. 1/PEP, fixed ADP-Mg | 1/v vs. 1/ADP-Mg, fixed PEP3- | 1/v vs. 1/PEP3-, fixed ADP-Mg | 1/v vs. 1/ ADP-Mg, fixed PEP3- | ||
| C | NC | MT | C | 0.05±0.002 | 35 ±1.2 |
Inhibition patterns were taken from the double reciprocal plots of the inhibition experiments (Fig. 3). Simple inhibition patterns were confirmed from linear replots of the slopes or intercepts versus the inhibitor concentrations (not shown). The inhibition constants were calculated from the fits of the complete data set to the corresponding equations for linear competitive inhibition (C) v = V[S]/(K m (1+ [I]/K i) +[S]), linear noncompetitive inhibition (NC), or linear mixed inhibition (MT) v = V[S]/(K m (1+ [I]/K i) + [S](1+[I]/αK i)), where α = 1 and α < 1 for NC and MT, respectively; K i is the inhibition constant.
Thermostability of TpPK analyzed by DSC
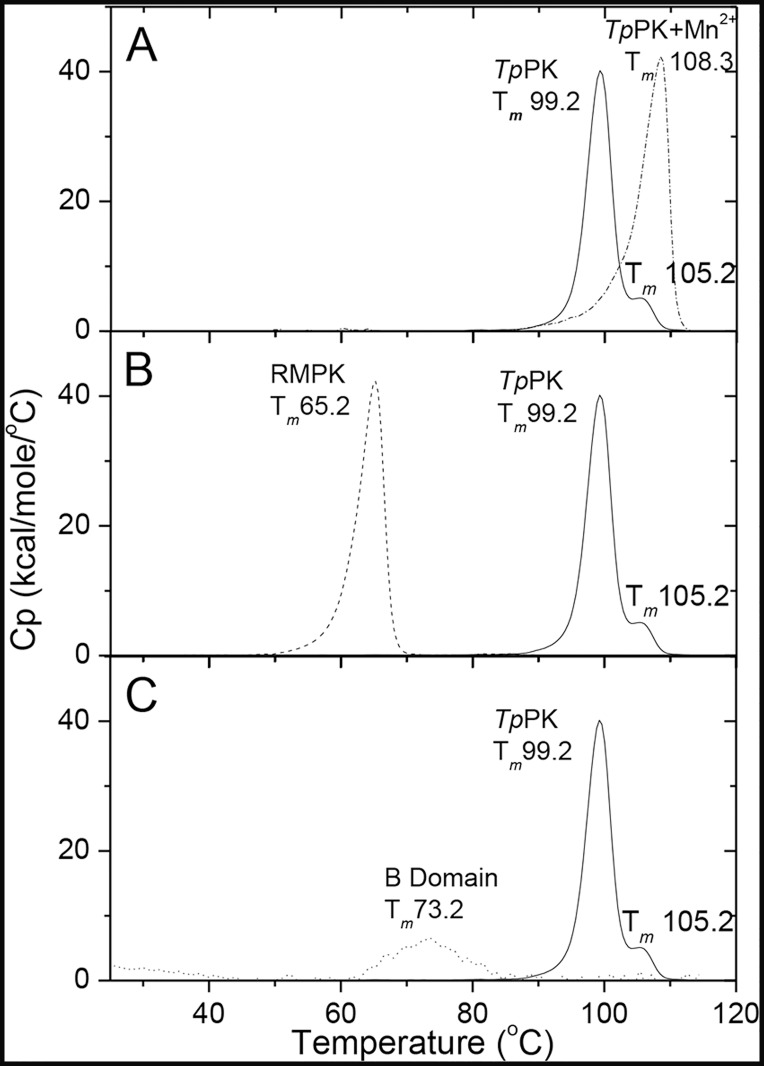
Because TpPK is a hyperthermophilic enzyme, we were interested in studying and comparing its thermal stability with the mesophilic RMPK. Thermal denaturation of TpPK was studied by DSC at a rate of 2.5°C/min at various protein concentrations in the range of 0.1–2.0 mg/ml. The T ms were similar and independent of the protein concentration within the 10- to 20-fold range. This result strongly suggests that the dissociation of the subunits is not likely to be involved in the denaturation process, and no evident aggregation was observed within this protein concentration range. In all cases, the calorimetric transitions were irreversible, as demonstrated by the lack of a thermal effect in reheating runs. In addition, these transitions were also strongly dependent on the scanning rates (within the range of 0.5–2.5°C/min), indicating that the denaturation process of TpPK is under kinetic control (data not shown). It is relevant to mention that under these experimental conditions, TpPK exhibited two calorimetric transitions. Additional DSC experiments were conducted at 1 mg/ml and 1.5°C/min. In Fig. 4A, the thermograms for TpPK with and without 0.2 mM MnCl2 are shown. The T m values of the two transitions of TpPK without Mn2+ were 99.2 and 105.2°C; with Mn2+, the T m value increased to 108.4°C, and a single transition was observed. This result indicated that the enzyme was stabilized with Mn2+ and that the denaturation occurred in a single step. It is known that metal ions that bind with high affinity to specific sites often stabilize the conformation of proteins [57,58].
Fig 4. Differential scanning calorimetry of the TpPK.
A. TpPK without and with 0.2 mM Mn2+ are represented by solid and dashed-dotted lines, respectively. B. TpPK: solid line, RMPK: dashed line, C. TpPK: solid line, the B domain: dotted line. The enzyme concentration was 1.0 mg/ml (19.47 μM monomer) for TpPK, the B domain concentration was 0.18 mg/ml (19.55 μM of domain). The scan rate was 1.5°C/min.
In Fig. 4B, the thermogram for the hyperthermophilic TpPK was compared with that of the mesophile RMPK. In contrast to the two calorimetric transitions of TpPK, RMPK only exhibited one transition with a T m of 65°C. The two transitions observed in the thermogram of TpPK suggested independent denaturation of its domains; whereas the thermogram of RMPK indicated a single global denaturation. Similar transitions were observed before or after removal of the His6 tag of recombinant TpPK, ruling out the possibility that any of the transitions were due to the extra peptide (data not shown).
To assess whether the second transition corresponded to the B domain, this domain was cloned, overexpressed and purified as described in the Materials and Methods section. Fig. 4C shows that the T m of the B domain was 73.5°C, i.e., very low compared to the second transition of TpPK (105.2°C). It seems therefore that inter-domain interactions of TpPK stabilize the B domain as reported for the β-1,4-glycanase from Cellulomonas fimi [59]. Therefore, the DSC of the isolated domain of TpPK is not sufficient to define the nature of the 105°C transition. However, when a single isolated β-sheet barrel domain is stable, a single transition coincident with the second transition of a whole protein has been reported [59].
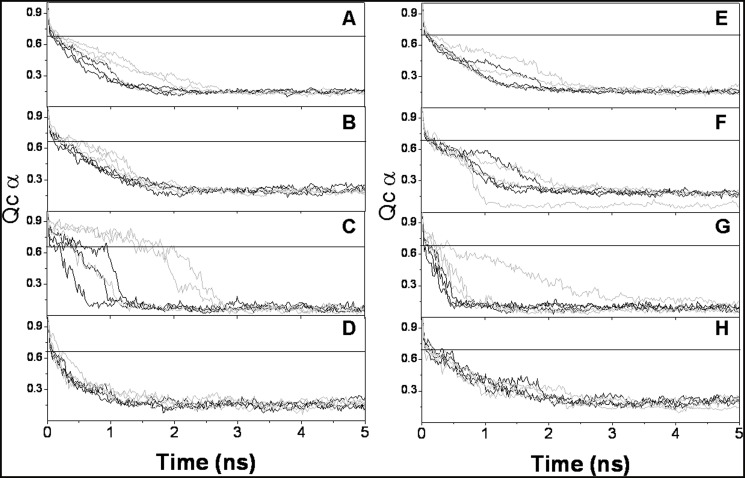
Because the second 105.2°C transition could not be matched with that of the isolated B domain of TpPK, this result could be attributed either to this domain or to any other rearrangement of the protein. Therefore, to gain insights on the origin of this second transition, a molecular dynamic simulation of a modeled monomer of TpPK was carried out. Evaluation of the model quality is shown in S1 Table. In Fig. 5A, the time course for the first 5 ns of the 50 ns for the triplicate molecular dynamics at 500 K are shown for the monomers of TpPK and RMPK. The Q values for TpPK and RMPK were calculated for the whole monomer and for the A, B and C domains. The analysis of the native contacts within each domain of TpPK vs. time indicated that the A and C domains lost 90% of their native contacts within 2–3 ns (Fig. 5B and 5D). In contrast, domain B retained a high Q value (~0.65) for up to 2.5 ns (Fig. 5C). It is noted that the most stable simulations of domain B of TpPK were those that started from the closed structure. However, even those that started from the open structure were more stable than the RMPK counterpart. For RMPK, only one simulation showed a B domain reaching Q = 0.2 at approximately 2.5 ns. The rest were denatured before 1 ns. For reference, a value of Q = 0.7 is defined as the limit before a 60-residue helical protein leaves the native state to approach the molten globule state [60]. Further analysis of the denaturation process is shown in S5 Fig., in which the loss of secondary structure can be seen. It is clear from the simulations that at 500 K, the proteins are completely denatured. These results suggest that the second calorimetric transition of TpPK is due to the thermal denaturation of the B domain, whereas RMPK was denatured in a single global event. Remarkably, TpPK simulated at 300 K exhibited closure of the B domain over the A domain. The same phenomenon occurred in the three simulations. Under the same conditions, two of the three simulations of RMPK completed with an open cleft and one ended with a twisted lid over the A domain (S1 File https://docs.google.com/file/d/0B57RfHIF-7vbNG5KZDg0N0pIS28/edit?usp=drive_web).
Fig 5. Loss of native contacts (QCα) versus time for the modeled TpPKs and RMPKs at 500 K.
The native contacts for the PK monomer and of the A, B and C domains are shown. TpPKs and RMPKs correspond to panels A through D and to panels E through H, respectively. Simulations were performed for 50 ns in an implicit solvent at 500 K. The simulations were started from two different conformations of the enzymes, open (black lines) and closed (grey lines). The open and closed conformations for TpPK were modeled as described in Material and Methods. The open and closed conformations for RMPK were obtained for PDBID 2G50 and PDBID 1A5U, respectively. The simulations were run in triplicate. Although the simulations were run for 10 to 50 ns, only the relevant periods for the transitions are shown.
Comparative analysis of the structure of a mesophilic and a hyperthermophilic PK
Mesophilic PKs exhibit a high mobility of the B domain and are stabilized when the active site is partially or totally occupied. In this respect, Larsen et al. [61,38] determined the structure of RMPK in the presence of various ligands. The authors found that changes in the position of the B domain relative to the rest of the protein account for the various conformations. When the active site cleft is closed, the angle of reference is 0° and the active site is completely occupied by the complex K+-Mg2+-oxalate-ATP-Mg. When the active site is partially occupied by K+, Mg2+ and an analog of PEP, the B domain can exhibit different angles of rotation (11°, 21° or 41°) relative to the closed subunits, indicating that the closure of the active site varies from partially closed to totally open [61,38]. In the crystal structure of the PK apoenzyme from cat muscle, no electronic density was found for the B domain [62]. This result suggests that when the active site is empty, the mobile B domain cannot be modeled due to weak or absent electron density.
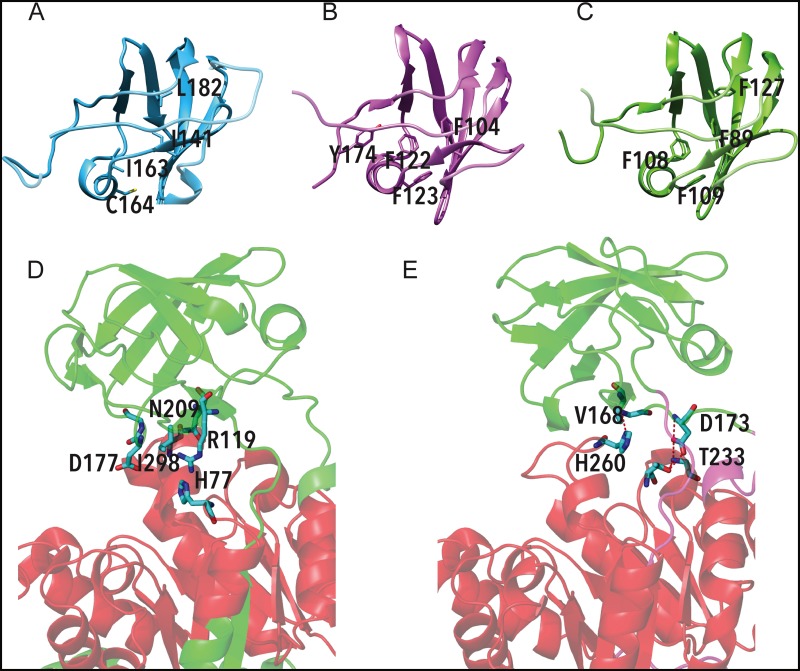
In contrast to the described cat muscle apo-PK structure, a complete electron density of the B domain of molecule A in the asymmetric unit was observed in the crystal structure of PaPK (PDBID 3QTG), even though no ligands were present in the active site. This raises the question of how apo-PaPK manages to conserve the complete electron density of the cleft in the absence of ligands that stabilize the B domain. To address this, the intra-domain and inter-domain interactions between the A and B domains of RMPK were compared with those of apo-PaPK. More salt- bridges and beta-bridges were found in PaPK than in RMPK (data not shown). The B domain of TpPK was modeled and compared to those of PaPK and RMPK (Fig. 6A- 6C). The hydrophobic core is formed by aromatic residues (Phe) in PaPK as well as in the TpPK model, which is in contrast to the aliphatic residues found in RMPK (Fig. 6B and 6C vs. 6A). Phe122, Phe123 and Phe104 of PaPK correspond to Phe108, Phe109 and Phe89 of TpPK and to Ile163, Cys164 and Ile141 of RMPK. It is worth mentioning that in Crenarchaeota, aromatic residues at these positions are highly conserved (positions 89, 108 and 109 are 85%, 88% and 100% conserved), whereas in RMPK, positions 163, 164 and 141 are substituted by aliphatic residues (S6 Fig.). The positions of the Phe residues observed in both PaPK and TpPK favor edge-to-face orientations of π-π electrostatic interactions (Table 4). To explore whether the π-π interactions of the B domain play a role in the closure of the lid over the A domain, the mutant F89I/F108I/F109C/F127L of TpPK was modeled. In this mutant, we replaced the Phe residues present in TpPK with those found in the corresponding positions of RMPK. Three simulations of this mutant were run at 300 K (S2 File https://drive.google.com/file/d/0B57RfHIF-7vbN1ZiQjNoS0RFbk0/view?usp=sharing). In contrast with the simulations of TpPK, the open lid remained in two of the three simulations of the mutant TpPK, while the third ended with a closed lid. Thus, the Phe-Phe interactions at this hydrophobic core likely contribute to the high stability and closed conformation of the B domain of TpPK [63]. The interactions between domains A and B of RMPK and PaPK are shown in 6D and 6E, respectively. Note that the interactions present in RMPK are different from those found in PaPK. The interactions found in PaPK are mostly in the hinges that join domains A and B, which allow a semi-closed cleft conformation, whereas RMPK does not have these interactions. Table 5 summarizes the atoms involved in the interactions and the distances between them. These two sets of distinct interactions, those within the B domain and those between domains A and B, may account for the putative high thermostability of the B domain of TpPK. The high stability and low mobility of the active site cleft of the hyperthermophilic enzyme suggest that the structural arrangements could allow the catalytic activity of TpPK in the absence of a positive charge. We are currently generating the constructs according to the structural analysis of the active site cleft to assess this working hypothesis.
Fig 6. Models of the hydrophobic core of the B domain of RMPK (A), PaPK (B) and TpPK (C). Interactions between domains A and B of RMPK (D) and PaPK (E).
The aromatic residues at the hydrophobic core are represented as sticks. The residues involved in polar and hydrophobic interactions between domain A and domain B are shown as sticks. The red dotted lines highlight polar interactions. Notice that these interactions are absent in RMPK. This figure was constructed from the coordinates of RMPK and PaPK deposited under file names 2G50 and 3QTG at the PDB. It is noteworthy that the interdomain interactions were not analyzed in the modeled monomer of TpPK but were determined from the structure of PaPK to obtain reliable results.
Table 4. π-π interactions between phenylalanine pairs in the hydrophobic core of the B domain of pyruvate kinases from Thermofilum pendens (modeled) and from Pyrobaculum aerophilum (PDB ID 3QTG).
| Interaction between residues | Distance (Å) | Angle (grades) | ||
|---|---|---|---|---|
| TpPK | F89 | F127 | 5.7 | 87.5 |
| F89 | F108 | 5.6 | 51.7 | |
| F108 | F109 | 5.0 | 87.7 | |
| PaPK | F104 | F122 | 5.9 | 32.7 |
| F122 | F123 | 5.1 | 87.7 | |
| F122 | Y174 | 6.4 | 55.1 | |
The distances were measured from the mass center, defined by the carbon atoms that form the aromatic rings. The angles were measured from the planes formed by the carbon atoms in each ring. Phe 89, 108 and 109 of TpPK correspond to Phe 104, 122 and 123 of PaPK and are 85%, 88% and 100% conserved in all Crenarchaeota phyla. Notice that position 109 in the enzyme from the Sulfolobales consists primarily of Tyr.
Table 5. Interactions between the A and B domains of RMPK (PDB ID 2G50) and of PaPK (PDB ID 3QTG).
| PDB | Donor | Acceptor | Distance (Å) | Contact type |
|---|---|---|---|---|
| 2G50 | I298-CG2 | N209-CG | 3.61 | Hydrophobic |
| I298-CG2 | N209-CB | 3.59 | Hydrophobic | |
| I298-CG2 | G178-CA | 3.88 | Hydrophobic | |
| I298-CD1 | D177-C | 3.52 | Hydrophobic | |
| I298-CD1 | D177-CA | 3.72 | Hydrophobic | |
| R119-CZ | H77-CE1 | 3.79 | Hydrophobic | |
| 3QTG | G235-N | D173-OD2 | 2.98 | Polar |
| T233-OG1 | D173-OD1 | 2.60 | Polar | |
| G171-N | H260-ND1 | 3.03 | Polar | |
| G235-N | D173-CG | 3.62 | Hydrophobic | |
| T233-CB | D173-CG | 3.82 | Hydrophobic | |
| H260-CE1 | V168-CG1 | 3.51 | Hydrophobic |
The hinge regions, as determined by DynDom, were excluded from the analysis.
Conclusions
TpPK is encoded by a single-copy gene that is closely related to sequences from members of the Crenarchaeotal order Thermoproteales. These sequences are clustered in a group that includes PKs from the orders Desulfurococcales, Acidilobales, and Fervidicoccales, which contain amino acids other than Lys in the position corresponding to 117. Remarkably, TpPK has Val70 at the corresponding position, and it does not require an internal positive charge near the active site for catalysis. Nevertheless, it follows a rapid-equilibrium random-order kinetic mechanism equal to that of RMPK in the presence of K+ and the E117K mutant in the absence of K+. These results indicate that in TpPK, the closure of the active site and the arrangement of the residues involved in the binding of the nucleotide are independent of the internal positive charge and K+. The interactions (salt bridges, beta bridges, H+-bonds, hydrophobic and π-π interactions) present in the B domain of PaPK and those modeled in TpPK were analyzed and compared to those present in RMPK. The different interactions suggest that the B domain of the hyperthermophilic enzyme is highly stable. Taken together, the results lead to the working hypothesis that the structural arrangement of TpPK allows catalysis by the enzyme in the absence of a positive charge.
Supporting Information
Unrooted phylogenetic tree that includes all available PK protein sequences from the Archaea domain. Selected sequences from the Bacteria and the Eukarya domains were included as outer groups. Branches are colored according to the taxonomic group they belong. Logos showing conservation of the residues 113, 114, 117, and 120 (according to RMPK numbering) are shown adjacent to each taxonomic group included in the tree. The tree was inferred from 500 replicates, using the Maximum Likelihood method (18). The best tree with the highest log likelihood (-193785.3848) is shown. Similar trees were obtained with maximum-parsimony, minimum-evolution and neighbour-joining methods. The analysis involved 426 amino acid sequences (204 from archaea, 151 from bacteria and 71 from eukarya). The branches in the unrooted tree are drawn to scale, with the bar length indicating the number of substitutions per site. The proportion of replicate trees in which the associated taxa clustered together in a bootstrap test (500 replicates) is given next to selected branches.
(DOCX)
In (A) M indicates low molecular weight markers. In (B) the RMPK was used as molecular weight marker.
(DOCX)
The reaction mixture contained 50 mM Tris-HCl pH 6.0, 0.2 mM NADH, 0.1 mM Mn2+ free, 3.0 mM ADP-Mn complex and 8 μg/ml LDH. In plot A the Ribose-5-phosphate concentrations were 0 (∎), 0.5 (●), 1 (▴) and 5 (▾). In plot B the fructose-1,6-bisphosphate concentrations were 0 (∎), 0.5 (●), 1 (▴), 5 (▾), and 10 mM (◆). The reaction was started by the addition of the PK. The amounts of the PK ranged from 0.15 to 1.2 μg/ml.
(DOCX)
The experimental conditions were those described in Fig. 2.
(DOCX)
. The structural stability at the indicated temperatures is shown. The B domain is highlighted by a green square.
(DOCX)
. The residues that comprise the hydrophobic core of the B domain, at position 89, 108, 109, and 127 (numbering according to the TpPK), and equivalent positions in animals (numbering according to the RMPK) are showed.
(DOCX)
The movie shows a 3 by 3 animation where the 3 RMPK simulations at 300 K are on the top while the 3 TpPK simulations at 300 K are at the bottom. All the simulations were run for 50 ns. The proteins are represented on the basis of the secondary structure and colored accordingly. PK480p.mov https://docs.google.com/file/d/0B57RfHIF-7vbNG5KZDg0N0pIS28/edit?usp = drive_web.
(DOCX)
The movie shows a triplicate animation of the mutant (F89I/F108I/F109C/F127L) of the TpPK at 300 K. All the simulations were run for 50 ns. The proteins are represented on the basis of the secondary structure and colored accordingly. PKmutlid1080.mov https://drive.google.com/file/d/0B57RfHIF-7vbN1ZiQjNoS0RFbk0/view?usp=sharing.
(DOCX)
(DOCX)
The data of S3A Fig. were fitted (nonlinear regression Origin version 6.0) to the Hill equation v = V max*[S]n/K 0.5 n+[S]n. The mean and standard deviation of five experiments are shown.
(DOCX)
. The data of S3B Fig. were fitted (nonlinear regression Origin version 6.0) to the Hill equation v = V max*[S]n/K 0.5 n+[S]n. The mean and standard deviation of five experiments are shown.
(DOCX)
Acknowledgments
It is with profound respect and admiration that we dedicate this article to the memory of Professor Armando Gómez-Puyou from the Instituto de Fisiología Celular, UNAM who devoted his entire life to scientific research and made outstanding contributions to scientific development in Mexico, particularly in the areas of bioenergetics and the physicochemistry of proteins. We thank Dr. Miguel Costas of the Laboratorio de Biofisicoquímica, Departamento de Fisicoquímica, Facultad de Química, UNAM, for the use of the VP-DSC equipment. Dr. Claudia Reich, University of Illinois at Urbana-Champaign) supplied the genomic DNA of Thermophilum pendens. Dr. Maire Christine Slommiary of Université des Sciences at Technologies de Lille, Villeneuve d′ Ascq, France performed the mass spectroscopy of the B domain of TpPK. Francisco Mendoza Hoffmann helped to perform the BN-PAGE.
We thank Dr. Marietta Tuena de Gómez-Puyou for her valuable suggestions, Dr. Armando Gómez-Puyou for kind help and valuable ideas, and Dr. Andrés Ramírez-Silva, Dr. Juan Pablo Pardo-Vázquez and Dr. Oscar Juárez for careful revision of the manuscript. Dr. Lenin Domínguez-Ramírez gratefully acknowledges the support of the NVIDIA Corporation for the donation of the Tesla K20 GPU used for this research. Gustavo de la Vega-Ruíz was supported by a CONACyT PhD fellowship, number 265039, and this work is part of his PhD thesis for Postgraduate Studies in Biochemical Sciences of UNAM (Doctorado en Ciencias Bioquímicas, MyDCB). This work was carried out at the School of Medicine “Facultad de Medicina” of UNAM with L. Ramírez-Silva as his PhD advisor.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
Dirección General de Apoyo al Personal Académico-Universidad Nacional Autónoma de México, Grants IN215912 (to L.R.-S.), IN216513 (to H. R.-R.), and Grant IN201213 (to A.T.-L.) and by Consejo Nacional de Ciencia y Tecnología Grant CB2011-164838 (to A.T.-L.). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Kachmar JF, Boyer PD. Kinetic analysis of enzyme reactions. II. The potassium activation and calcium inhibition of pyruvic phosphoferase. J Biol Chem. 1953; 200: 669–682. [PubMed] [Google Scholar]
- 2. Mildvan AS, Cohn M. Kinetics and magnetic resonance studies of the pyruvate kinase reaction. II Complexes of enzyme, metal, and substrates. J Biol Chem. 1966; 24: 1178–1193. [PubMed] [Google Scholar]
- 3. Kayne FJ, Reuben J. Thallium (I) activation of pyruvate kinase. J Am Chem Soc. 1970; 92: 220–222. [Google Scholar]
- 4. Reuben J, Kayne FJ. Thallium-205 nuclear magnetic resonance study of pyruvate kinase and its substrates. J Biol Chem. 1971; 246: 6227–6234. [PubMed] [Google Scholar]
- 5. Nowak T, Mildvan AS. Nuclear magnetic resonance studies of the function of potassium in the mechanism of pyruvate kinase. Biochemistry. 1972; 11: 2819–2828. [DOI] [PubMed] [Google Scholar]
- 6. Raushel FM, Villafranca JJ. A multinuclear nuclear magnetic resonance study of the monovalent-divalent cation sites of pyruvate kinase. Biochemistry. 1980;19: 5481–5485. [DOI] [PubMed] [Google Scholar]
- 7. Oria-Hernández J, Cabrera N, Pérez-Montfort R, Ramírez-Silva L. Pyruvate kinase revisited. The activating effect of K+ . J Biol Chem. 2005; 280: 37294–37929. [DOI] [PubMed] [Google Scholar]
- 8. Oria-Hernández J, Riveros-Rosas H, Ramírez-Silva L. Dichotomic phylogenetic tree of the pyruvate kinase family. K+-dependent and independent enzymes. J Biol Chem. 2006; 281: 30717–30724. [DOI] [PubMed] [Google Scholar]
- 9. Laughlin IT, Reed G H. The monovalent cation requirement of rabbit muscle pyruvate kinase is eliminated by substitution of lysine for glutamate 117. Arch Biochem Biophys. 1997; 348: 262–267. [DOI] [PubMed] [Google Scholar]
- 10. Schramm A, Siebers B, Tjaden B, Brinkmann H, Hensel R. Pyruvate kinase of th hyperthermophilic crenarchaeota Thermoproteus tenax: physiological role and phylogenetic aspects. J Bacteriol. 2000; 182: 2001–2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Johnsen U, Hansen T, Schonheit P. Comparative analysis of pyruvate kinases from the hyperthormophilic archaea Archaeoglobus fulgidus, Aeropyrum pernix and Pyrobaculum aerophilum and the hyperthermophilic bacterium Thermotoga maritima: unusual regulatory properties in hyperthermophilic archaea. J Biol Chem. 2003; 278: 25417–25427. [DOI] [PubMed] [Google Scholar]
- 12. UniProt Consortium. Activities at the universal protein resource (UniProt). Nucleic Acids Res. 2014;42: D191–D198. 10.1093/nar/gkt1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007; 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 14. Madej T, Lanczycki CJ, Zhang D, Thiessen PA, Geer RC, Marchler-Bauer A, et al. MMDB and VAST+: tracking structural similarities between macromolecular complexes. Nucleic Acids Res. 2013; 42: D297–D303. 10.1093/nar/gkt1208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rose PW, Bi C, Bluhm WF, Christie CH, Dimitropoulos D, Dutta S, et al. The RCSB protein data bank: new resources for research and education. Nucleic Acids Res. 2013; 41: D475–D482. 10.1093/nar/gks1200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser. 1999; 41: 95–98. [Google Scholar]
- 17. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011; 28: 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Whelan S, Goldman N. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol. 2001; 18: 691–699. [DOI] [PubMed] [Google Scholar]
- 19. Posada D., and Buckley T. R. Model selection and model averaging in phylogenetics: Advantages of Akaike information criterion and Bayesian approaches over likelihood ratio tests. Syst Biol. 2004; 53: 793–808. [DOI] [PubMed] [Google Scholar]
- 20. Crooks GE, Hon G, Chandonia J M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004; 14: 1188–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Stols L, Gu M, Dieckman L, Raffen R, Collart FR, Donelly MI. A new vector for high-throughput, ligation-independent cloning encoding a tobacco etch virus protease cleavage site. Protein Expr Purif. 2002; 25: 8–15. [DOI] [PubMed] [Google Scholar]
- 22. Schägger H, von Jagow G. Blue native electrophoresis for isolation of membrane protein complexes in enzymatically active form. Anal Biochem. 1991; 199: 223–231. [DOI] [PubMed] [Google Scholar]
- 23. Kasahara M, Penefsky HS. High affinity binding of monovalent Pi by beef heart mitochondrial adenosine triphosphatase. J Biol Chem. 1978; 253: 4180–4187. [PubMed] [Google Scholar]
- 24. Ramírez-Silva L, de Gómez-Puyou MT, Gómez-Puyou A. Water-induced transitions in the K+ requirements for the activity of pyruvate kinase entrapped in reverse micelles. Biochemistry. 1993; 32: 5332–5338. [DOI] [PubMed] [Google Scholar]
- 25. Büchner T, Pleiderer G. Pyruvate kinase from muscle, in Methods in Enzymol (Colowick S. and Kaplan N. Eds.) 1955; vol. 1: 435–440. Academic Press, New York. [Google Scholar]
- 26. Schoemakers JM, Visser GJ, Flik G, Theuvenet PR. CHELATOR: an improved method for computing metal ion concentrations in physiological solutions. Biotechniques. 1992; 12: 870–879. [PubMed] [Google Scholar]
- 27. Susan-Resiga D, Nowak T. Proton donor in yeast pyruvate kinase: chemical and kinetic properties of the active site Thr298 to Cys mutant. Biochemistry. 2004; 43: 15230–15245. [DOI] [PubMed] [Google Scholar]
- 28. Dougherty TM, Cleland WW. pH Studies of the chemical mechanism of rabbit muscle pyruvate kinase. 2. Physiological substrates and phosphoenol-αketobutyrate. Biochemistry. 1985; 24: 5875–5880. [DOI] [PubMed] [Google Scholar]
- 29. Eswar N, Marti-Renom MA, Webb B, Madhusudhan MS, Eramian D, Shen M, et al. Comparative Protein Structure Modeling With MODELLER. Current Protocols in Bioinformatics, John Wiley & Sons, Inc., Supplement. 2006; 15: 5.6.1–5.6.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Solomons TG, Johnsen U, Schönheit P, Davies C.3-Phosphoglycerate is an Allosteric Activator of Pyruvate Kinase from the Hyperthermophilic Archaeon Pyrobaculum aerophilum . Biochemistry. 2013; 52: 5865–5875. 10.1021/bi400761b [DOI] [PubMed] [Google Scholar]
- 31. Chen VB, Arendall WB III, Headd JJ, Keedy DA, Immormino RM, Kappal GJ, et al. Molprobity: All atom structure validation for macromolecular crystallography. Acta Crystallographica. 2010; D66:12–21. 10.1107/S1744309109042018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera- a visualization system for exploratory research and analysis J Comput Chem. 2004; 25: 1605–1612. [DOI] [PubMed] [Google Scholar]
- 33. Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1996; 8: 127–134. [DOI] [PubMed] [Google Scholar]
- 34. Case DA, Darden TA, Cheatham TE III, Simmerling CL, Wang J, Duke RE, et al. AMBER 12. University of California, San Francisco; 2012. [Google Scholar]
- 35. Goetz AW, Williamson MJ, Xu D, Poole D, Le Grand S, Walker RC. Routine microsecond molecular dynamics simulations with AMBER on GPUs. 1. Generalized Born. J Chem Theory Comput. 2012; 8: 1542–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Glykos NM. Carma: a molecular dynamics analysis program J Comput Chem. 2006; 27: 1765–1768. [DOI] [PubMed] [Google Scholar]
- 37. Koukos PI, Glykos NM. grcarma: A Fully Automated Task-Oriented Interface for the Analysis of Molecular Dynamics Trajectories, J Comput Chem. 2013; 34: 2310–2312. [DOI] [PubMed] [Google Scholar]
- 38. Larsen TM, Benning MM, Rayment I, Reed GH. Structure of the bis(Mg2+)- ATP-oxalate complex of the rabbit muscle pyruvate kinase at 2.1 Å resolution: ATP binding over a barrel. Biochemistry. 1998; 37: 6247–6255. [DOI] [PubMed] [Google Scholar]
- 39. Muñoz ME, Ponce E. Pyruvate kinase: current status of regulatory and functional properties. Comp Biochem Physiol Part B. 2003; 135:197–218. [DOI] [PubMed] [Google Scholar]
- 40. Liao CL, Atkinson DE. Regulation at the phosphoenolpyruvate branchpoint in Azotobacter vinelandii: pyruvate kinases. J Bacteriol. 1971; 106: 37–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Smart JB, Pritchard GG. Regulation of pyruvate kinase from Propionibacterium shermanii . J Gen Microbiol. 1979; 122: 281–288. [DOI] [PubMed] [Google Scholar]
- 42. Kapoor R, Venkitasubramanian TA. Glucose 6-phosphate activation of pyruvate kinase from Mycobacterium smegmatis . Biochem J. 1981; 193: 435–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Malcovati M, Valentini G. AMP- and fructose 1,6-bisphosphate activated pyruvate kinases from Escherichia coli. Methods Enzymol. 1982; 90: 170–179. [DOI] [PubMed] [Google Scholar]
- 44. Busto F, Del Valle P, Soler J. Some kinetic properties of pyruvate kinase from Phycomyces blakesleeanus . Biochem Cell Biol. 1988; 66: 148–157. [Google Scholar]
- 45. Jetten SM, Gubler ME, Lee SH, Sinskey AJ. Structural and funtional analysis of pyruvate kinase from Corynebacterium glutamicum . Appl Environ Microbiol. 1994; 60: 2501–2507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Steiner P, Fussenegger M, Bailey JE, Sauer U. Cloning and expression of the Zymomonas mobilis pyruvate kinase gene in Escherichia coli. Gene. 1988; 220: 31–38. [DOI] [PubMed] [Google Scholar]
- 47. Mildvan AS, Cohn M. Kinetic and magnetic resonance studies of the pyruvate kinase reaction. J Biol Chem. 1965; 240: 238–246. [PubMed] [Google Scholar]
- 48. Dupont CL, Butcher A, Valas RE, Bourne PE, Caetano-Anallés G. History of biological metal utilization inferred through phylogenomic analysis of protein structures. Proc Natl Acad Sci USA. 2010. 107: 10567–10572. 10.1073/pnas.0912491107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Reynard AM, Hass LF, Jacobsen DD, Boyer PD. The correlation of reaction kinetics and substrate binding with the mechanism of pyruvate kinase. J Biol Chem. 1961; 236: 2277–2283. [PubMed] [Google Scholar]
- 50. Mildvan AS, Cohn M. Kinetics and magnetic resonance studies of the pyruvate kinase reaction. II Complexes of enzyme, metal, and substrates. J Biol Chem. 1966; 241: 1178–1193. [PubMed] [Google Scholar]
- 51. Ainsworth S, MacFarlane N. A kinetic study of rabbit muscle pyruvate kinase. Biochem J. 1973; 131: 223–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Cleland WW.in The Enzymes, Vol. 2, 3rd Ed (Boyer P. D., ed). 1970: 1–65, Academic Press, Inc., New York. [Google Scholar]
- 53. Reed GH, Morgan SD. Kinetic and magnetic resonance studies of the interaction of oxalate with pyruvate kinase. Biochemistry. 1974; 13: 3537–3541. [DOI] [PubMed] [Google Scholar]
- 54. Gupta RK, Benovic JL. Magnetic resonance and kinetic studies of the spatial arrangement of phosphoenolpyruvate and chromium (III) adenosine diphosphate at the catalytic site of pyruvate kinase. J Biol Chem. 1978; 253: 8878–8896. [PubMed] [Google Scholar]
- 55. Ramírez-Silva L, Ferreira ST, Nowak T, Tuena de Gómez-Puyou M, Gómez-Puyou A. Dimethylsulfoxide promotes K+-independent activity of pyruvate kinase and the acquisition of the active catalytic conformation. Eur J Biochem. 2001; 268: 3267–3274. [DOI] [PubMed] [Google Scholar]
- 56. Ramírez-Silva L, Oria-Hernández J. Selectivity of pyruvate kinase for Na+ and K+ in water/dimethylsulfoxide mixtures. Eur J Biochem. 2003; 270: 2377–2385. [DOI] [PubMed] [Google Scholar]
- 57. Doolittle RF. The multiplicity of domains in proteins. Annu Rev Biochem. 1995; 64: 287–314. [DOI] [PubMed] [Google Scholar]
- 58. Ruiz-Arribas A, Santamaría RI, Zhadan GG, Villar E, Shnyrov VL. Differential scanning calorimetric study of the thermal stability of xylanase from Streptomyces halstedii JM8. Biochemistry. 1994; 33: 13787–13791. [DOI] [PubMed] [Google Scholar]
- 59. Nikolova PV, Creagh AL, Duff SJ, Haynes CA. Thermostability and Irreversible activity loss of exoglucanase/xylanase Cex from Cellulomonas fimi. Biochemistry. 1997; 36: 1381–1388. [DOI] [PubMed] [Google Scholar]
- 60. Onuchic JN, Wolynes PG, Luthey-Schulten Z, Socci ND. Toward an outline of the topography of a realistic protein-folding funnel Proc Natl Acad Sci USA. 1995; 92: 3626–3630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Larsen TM, Benning MM, Wesenberg GE, Rayment I, Reed GH. Ligand-induced domain movement in pyruvate kinase: structure of the enzyme from rabbit muscle with Mg2+, K+, and L-phospholactate at 2.7 Å resolution. Arch Biochem Biophys. 1997; 345: 199–206. [DOI] [PubMed] [Google Scholar]
- 62. Allen SC, Muirhead H. Refined three-dimensional structure of cat muscle (M1) pyruvate kinase at a resolution of 2.6 Å. Acta Crystallogr D Biol Crystallogr. 1996; 52: 499–504. [DOI] [PubMed] [Google Scholar]
- 63. Hunter CA, Singh J, Thornton JM. π-π Interactions: the geometry and energetics of phenylalanine-phenylalanine interactions in proteins. J Mol Biol. 1991. 218: 837–846. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Unrooted phylogenetic tree that includes all available PK protein sequences from the Archaea domain. Selected sequences from the Bacteria and the Eukarya domains were included as outer groups. Branches are colored according to the taxonomic group they belong. Logos showing conservation of the residues 113, 114, 117, and 120 (according to RMPK numbering) are shown adjacent to each taxonomic group included in the tree. The tree was inferred from 500 replicates, using the Maximum Likelihood method (18). The best tree with the highest log likelihood (-193785.3848) is shown. Similar trees were obtained with maximum-parsimony, minimum-evolution and neighbour-joining methods. The analysis involved 426 amino acid sequences (204 from archaea, 151 from bacteria and 71 from eukarya). The branches in the unrooted tree are drawn to scale, with the bar length indicating the number of substitutions per site. The proportion of replicate trees in which the associated taxa clustered together in a bootstrap test (500 replicates) is given next to selected branches.
(DOCX)
In (A) M indicates low molecular weight markers. In (B) the RMPK was used as molecular weight marker.
(DOCX)
The reaction mixture contained 50 mM Tris-HCl pH 6.0, 0.2 mM NADH, 0.1 mM Mn2+ free, 3.0 mM ADP-Mn complex and 8 μg/ml LDH. In plot A the Ribose-5-phosphate concentrations were 0 (∎), 0.5 (●), 1 (▴) and 5 (▾). In plot B the fructose-1,6-bisphosphate concentrations were 0 (∎), 0.5 (●), 1 (▴), 5 (▾), and 10 mM (◆). The reaction was started by the addition of the PK. The amounts of the PK ranged from 0.15 to 1.2 μg/ml.
(DOCX)
The experimental conditions were those described in Fig. 2.
(DOCX)
. The structural stability at the indicated temperatures is shown. The B domain is highlighted by a green square.
(DOCX)
. The residues that comprise the hydrophobic core of the B domain, at position 89, 108, 109, and 127 (numbering according to the TpPK), and equivalent positions in animals (numbering according to the RMPK) are showed.
(DOCX)
The movie shows a 3 by 3 animation where the 3 RMPK simulations at 300 K are on the top while the 3 TpPK simulations at 300 K are at the bottom. All the simulations were run for 50 ns. The proteins are represented on the basis of the secondary structure and colored accordingly. PK480p.mov https://docs.google.com/file/d/0B57RfHIF-7vbNG5KZDg0N0pIS28/edit?usp = drive_web.
(DOCX)
The movie shows a triplicate animation of the mutant (F89I/F108I/F109C/F127L) of the TpPK at 300 K. All the simulations were run for 50 ns. The proteins are represented on the basis of the secondary structure and colored accordingly. PKmutlid1080.mov https://drive.google.com/file/d/0B57RfHIF-7vbN1ZiQjNoS0RFbk0/view?usp=sharing.
(DOCX)
(DOCX)
The data of S3A Fig. were fitted (nonlinear regression Origin version 6.0) to the Hill equation v = V max*[S]n/K 0.5 n+[S]n. The mean and standard deviation of five experiments are shown.
(DOCX)
. The data of S3B Fig. were fitted (nonlinear regression Origin version 6.0) to the Hill equation v = V max*[S]n/K 0.5 n+[S]n. The mean and standard deviation of five experiments are shown.
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.