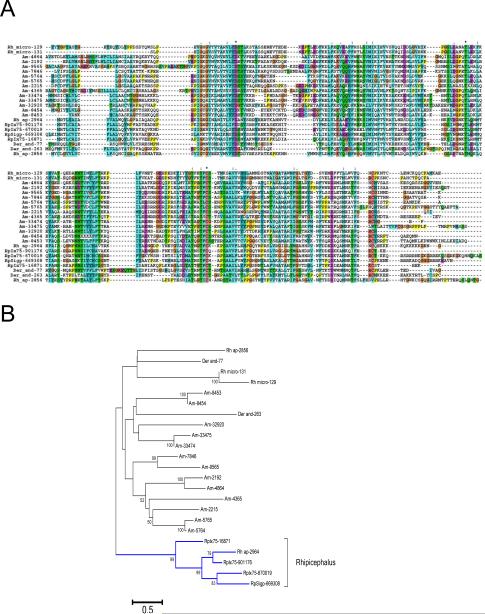
Figure 4. Novel metastriate tick salivary family.
(A) Clustal alignment. The symbols above the alignment indicates for * identity of residues, “:” similarity and “.” indicates less similarity. (B) Phylogenetic evolutionary history was inferred using the Neighbor-Joining method [134].The bootstrap consensus tree inferred from 10000. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates are collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (10000 replicates). All ambiguous positions were removed for each sequence pair. There were a total of 360 positions in the final dataset. The bar at the bottom indicates 50% amino acid divergence. Other conditions as in figure 1. The sequences were obtained from previous publications [2, 19]. The R. pulchellus sequences start with Rp and can be obtained from supplemental file 1.