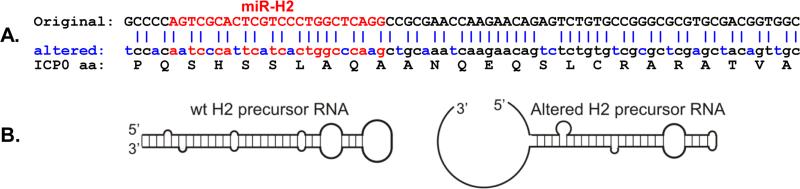
Fig. 2.
Altered codon usage of the ICP0 ORF region corresponding to the miR-H2 precursor RNA. A. Sequences are for the ICP0 region complementary to the H2 precursor miRNA. Changed nts are blue. Nts complementary to the H2 miRNA are red. H2 miRNA precursor: 21 of 75 nts (28%) are changed. H2 miRNA: 7 of 24 nts (29%) are changed. Seed sequence: 2 of 7 nts are changed. The predicted ICP0 amino acid sequence shown immediately under the “altered” nucleotide sequence is identical for the “original” (wt) and the “altered” (mutated) nucleotide sequences. B. The altered sequence changes the predicted structure of the RNA made from the complementary LAT DNA strand. Virgo software version 2.0 no longer predicts a miRNA precursor in this location. Thus, no H2 miRNA will be made. The amino acid sequence