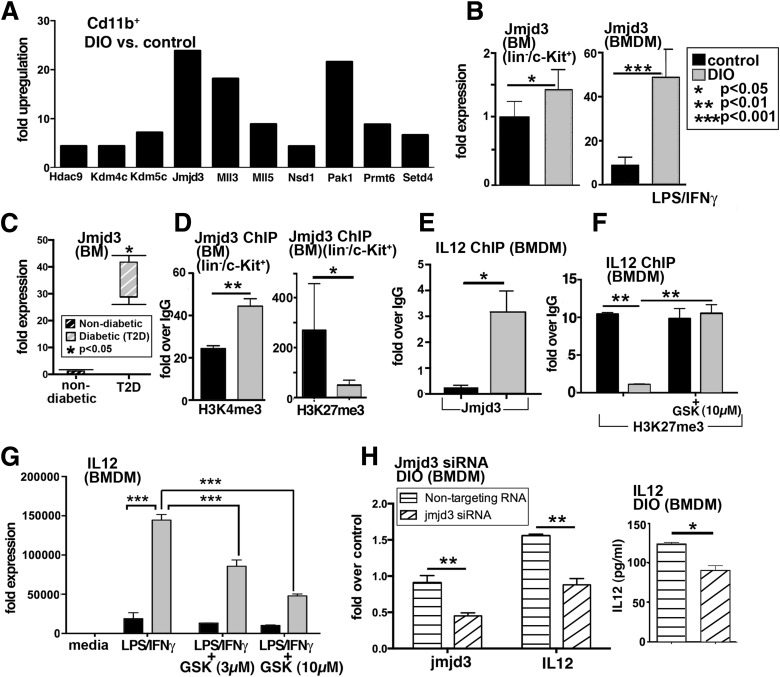
Figure 5.
Increased JmjC demethylase, Jmjd3, production in BM LK cells and macrophages. A: Gene expression of chromatin-modifying enzymes was analyzed using PCR array plates in CD11b+ MACS-isolated cells from DIO and control wounds at day 3. Enzymes that were upregulated are shown. The threshold was set to a fourfold difference. Data are expressed as fold over control (n = 3/group). B: RT-PCR quantification of Jmjd3 levels in DIO and control BM progenitor cells (lin−/c-Kit+) (isolated by MACS) and BMDMs (following stimulation with LPS/IFN-γ) (n = 4/group). C: RT-PCR analysis of human BM isolated from nondiabetic and T2D patients undergoing amputations at the femur level (n = 2/group). D: ChIP analysis of H3K4me3 and H3K27me3 on the Jmjd3 promoter of in vivo MACS-isolated BM LK cells from DIO and control mice (n = 5/group). E: ChIP analysis for Jmjd3 on the IL-12 promoter in DIO BMDMs compared with controls (n = 3/group, replicated once). F: BMDMs from DIO and control mice were treated with the H3K27me3 demethylase inhibitor, GSK-J4 (10 µmol/L), for 6 h, and ChIP analysis of H3K27me3 on the IL-12 promoter was performed (n = 3/group). G: BMDMs from DIO and control mice were stimulated with LPS/IFN-γ in the presence or absence of GSK-J4 (3 µmol/L, 10 μmol/L) for 6 h, and analysis of IL-12 transcription was performed (n = 3/group). Data are expressed as mean ± SE. H: DIO BMDMs were transfected with Jmjd3 siRNA or nontargeting siRNA or lipofectamine alone in triplicate wells. Forty-eight hours later, cells were stimulated with LPS/IFN-γ for 24 h. Transcript levels of Jmjd3 and IL-12 in cells treated with Jmjd3 siRNA or nontargeting siRNA are shown. Following stimulation, cell supernatants were collected and used for IL-12 Bioplex. Results are expressed as mean ± SE. Statistical analysis was performed using Student t tests, and P value <0.05 was considered significant.