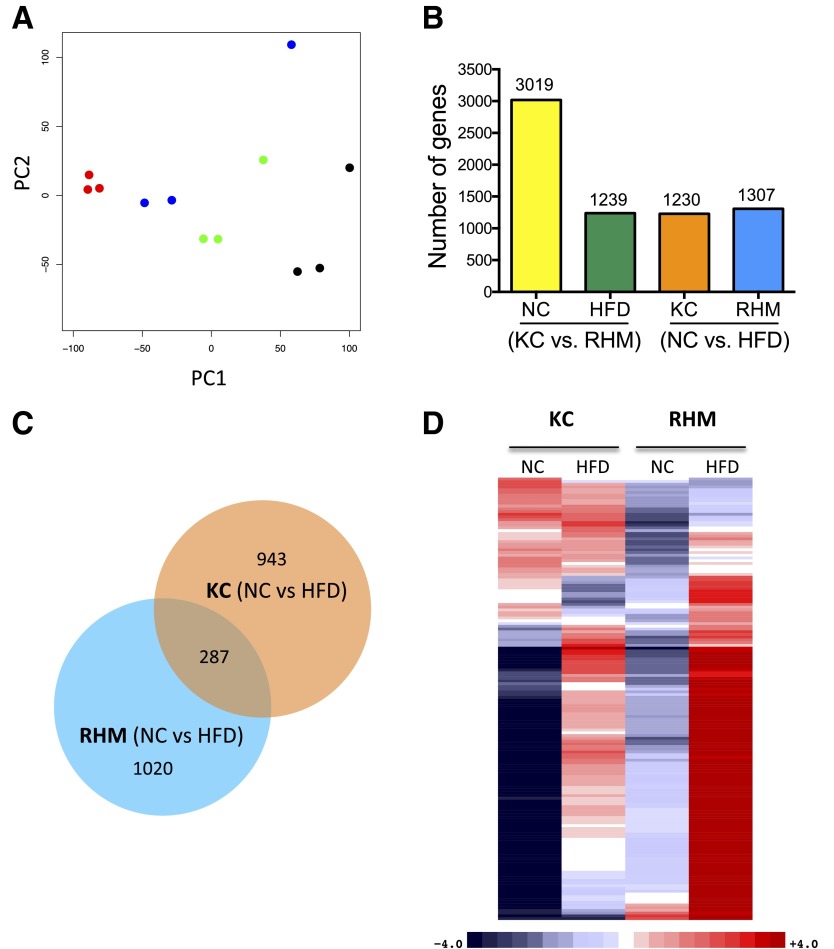
Figure 4.
RNA-seq analysis of KCs and RHMs from NC- and HFD-fed mice. A: Principal component analysis of expressed genes in KCs and RHMs from NC- or HFD-fed mice. Samples representing three biological replicates cluster together (KC-NC, red; KC-HFD, blue; RHM-NC, green; RHM-HFD, black). B: Bar chart shows the number of differentially expressed genes between cell types (KC vs. RHM) and diets (NC vs. HFD). C: Venn diagram shows common and distinct differentially expressed genes induced by the HFD within cell types. D: Heat map shows differentially expressed inflammatory response genes between KCs and RHMs from mice fed NC and the HFD in a four-way comparison. Gene counts were adjusted for cell number as KC increase 1.17-fold on HFD and RHMs increase 5.8-fold. Upregulated genes are colored red, downregulated genes are colored blue, and darker colors reflect higher fold changes. RNA-seq studies used three biological replicates per each group.