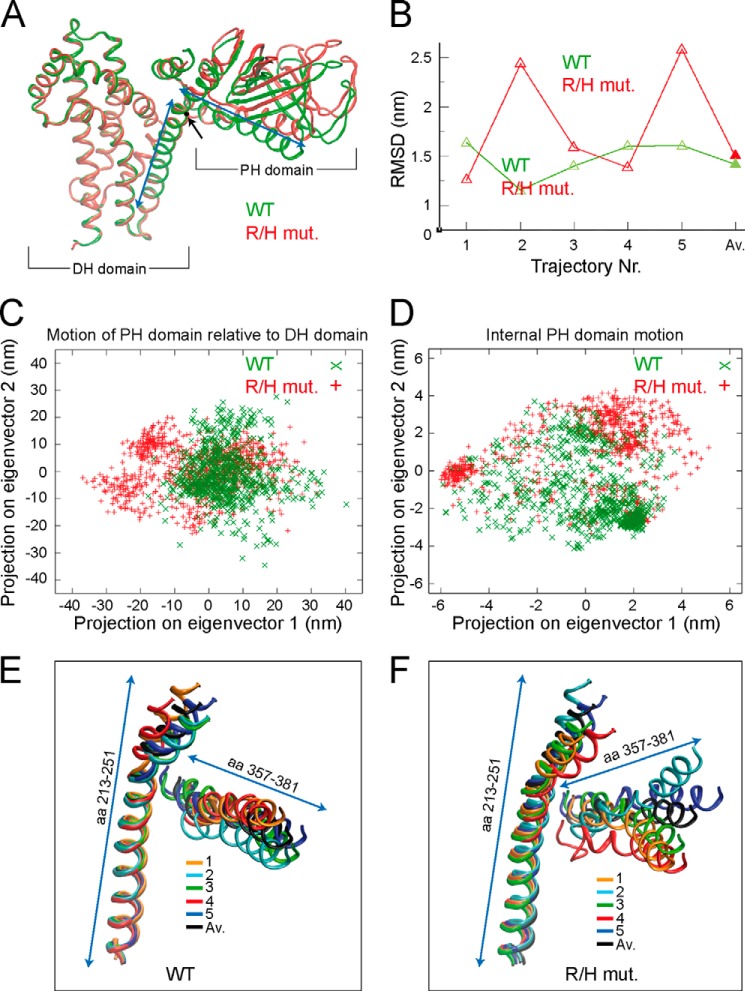
FIGURE 5.
The R/H mutation affects the orientation of the PH domain relative to the DH domain. A, ribbon backbone presentation of the superimposed average structures over five independent simulations of ΔSH3CbII WT (green) and the corresponding R/H mutant (red). For simplicity, the ribbon backbone of Cdc42 is omitted. The ΔSH3CbII DH and PH domains are indicated. The black dot and arrow indicate the position of the R/H mutation within the DH domain α-helix (left blue arrowed line). The right blue arrowed line indicates the neighboring PH domain α-helix. Note that the superimposed DH domains perfectly overlap, whereas the average PH domain of the R/H mutant shows a different orientation relative to the DH domain, as compared with WT. B, root mean square deviation (RMSD; WT, green; R/H mutant, red) of the whole PH domain backbone to the starting structure using the DH domain as the fitting group. Average structures of trajectories 1–5 (open triangles) and the average structures thereof (Av., closed triangles) are indicated. C, two-dimensional projections of the concatenated WT and R/H mutant trajectories, respectively, on the first and second eigenvector of the PH domain motion relative to the DH domain. D, two-dimensional projections on the first and second eigenvector of the internal PH domain motion. E, ribbon backbone presentation of the DH/PH α-helix (aa 213–251) and the PH domain α-helix (aa 357–381) of the average structures of the five independent ΔSH3CbII WT trajectories (colored) and the average structure thereof (black). F, Ribbon backbone presentation of the DH/PH α-helix (aa 213–251) and the PH domain α-helix (aa 357–381) of the average structures of the five independent ΔSH3CbII R/H mutant trajectories (colored) and the average structure thereof (black).