Background: M. tuberculosis two-component systems converge to co-regulate gene expression during nitrate metabolism.
Results: Analysis of nitrate/nitrite-responsive gene expression reveals novel interaction between NarL and DevR response regulators.
Conclusion: Cooperative binding of NarL and DevR enables co-regulation of a subset of genes.
Significance: The findings establish endogenous nitrite as a signal and highlight interplay between M. tuberculosis NarLS and DevRS/DosT signaling systems.
Keywords: Bacterial Signal Transduction, Gene Expression, Mycobacterium tuberculosis, Nitrogen Metabolism, Protein-Protein Interaction, Coregulation, DevR, NarL, Response Regulators, Two-component System
Abstract
Mycobacterium tuberculosis genes Rv0844c/Rv0845 encoding the NarL response regulator and NarS histidine kinase are hypothesized to constitute a two-component system involved in the regulation of nitrate metabolism. However, there is no experimental evidence to support this. In this study, we established M. tuberculosis NarL/NarS as a functional two-component system and identified His241 and Asp61 as conserved phosphorylation sites in NarS and NarL, respectively. Transcriptional profiling between M. tuberculosis H37Rv and a ΔnarL mutant strain during exponential growth in broth cultures with or without nitrate defined an ∼30-gene NarL regulon that exhibited significant overlap with DevR-regulated genes, thereby implicating a role for the DevR response regulator in the regulation of nitrate metabolism. Notably, expression analysis of a subset of genes common to NarL and DevR regulons in M. tuberculosis ΔdevR, ΔdevSΔdosT, and ΔnarL mutant strains revealed that in response to nitrite produced during aerobic nitrate metabolism, the DevRS/DosT regulatory system plays a primary role that is augmented by NarL. Specifically, NarL itself was unable to bind to the narK2, acg, and Rv3130c promoters in phosphorylated or unphosphorylated form; however, its interaction with DevR∼P resulted in cooperative binding, thereby enabling co-regulation of these genes. These findings support the role of physiologically derived nitrite as a metabolic signal in mycobacteria. We propose NarL-DevR binding, possibly as a heterodimer, as a novel mechanism for co-regulation of gene expression by the DevRS/DosT and NarL/NarS regulatory systems.
Introduction
Despite the availability of a vaccine and cost-effective antibiotics, tuberculosis continues to be a global health emergency. The remarkable ability of Mycobacterium tuberculosis to survive in nutrient-limited environments inside human macrophages and to persist for long periods of time necessitates studies to understand how M. tuberculosis coordinates adaptive responses that regulate growth, metabolism, and persistence.
Emerging evidence has revealed that highly integrated mycobacterial signaling networks play key roles in facilitating host-pathogen interactions and enabling intracellular survival strategies (1). The M. tuberculosis genome encodes 11 paired two-component signal transduction systems (TCSSs),6 two orphan histidine kinases (HKs), and 6 orphan response regulators (RRs) (2). Notably, two RRs (MtrA and PrrA) are essential for intracellular survival of M. tuberculosis (3, 4), and several others (DevR, PhoP, and MprA) have been implicated in virulence and persistence (reviewed in Ref. 5). Of all of the systems, the DevRS/DosT (also called DosRS/DosT) system is the most well characterized and regulates a ∼50-member regulon in response to hypoxia, nitric oxide (NO), carbon monoxide (CO), and ascorbic acid (6–13). More recently, studies have demonstrated a role for the DevR regulon in metabolic adaptation that is essential for M. tuberculosis to transition from an actively respiring to a latent non-replicating persistent state (14).
Nitrate holds a significant place in mycobacterial pathobiology, both for its role as an alternate terminal acceptor during anaerobic respiration presumed to occur as the bacilli enter a state of quiescence or dormancy (15–19) and for serving as a nitrogen source (20). It is well established that nitrate reduction to nitrite affords protection to hypoxic bacilli from reactive nitrogen and acid stresses encountered in vivo (21, 22). On the other hand, nitrate metabolism during aerobic growth of mycobacteria has just begun to be appreciated. It has recently been shown that M. tuberculosis-infected human macrophages produce large amounts of nitrite in vitro even at physiological oxygen tensions (23). These observations, together with the fact that nitrate is readily available from diet and from spontaneous degradation of NO (24), raise the possibility of nitrate assimilation. Although a nitrate assimilatory pathway in M. tuberculosis has recently been validated (25), its in vivo relevance is currently unknown. In any case, nitrite produced via nitrate reduction is a toxic by-product and is primarily secreted out of the bacilli by nitrite extrusion proteins. M. tuberculosis has three genes encoding for nitrate/nitrite transporter proteins (26), of which only one (narK2) has been extensively studied because of its induction by the DevRS/DosT TCSS (13) and implications in anaerobic respiration and latency. Despite the important role of nitrate in mycobacterial metabolism and respiration, its link to latency (27), and the underlying signaling interactions, the regulation of nitrate metabolism is still largely unknown.
In Escherichia coli, equilibrium between nitrate and nitrite concentrations regulate the activity of the NarX/NarQ HKs and NarL/NarP RRs (28). Based on the significant amino acid similarity to the NarL and NarP/NarL regulatory proteins of Pseudomonas aeruginosa and E. coli, respectively, the M. tuberculosis Rv0844c protein is annotated as NarL. In the present study, we investigated the role of M. tuberculosis NarL in the regulation of nitrate metabolism during aerobic growth conditions. We also established Rv0844c (narL)/Rv0845 (narS) genes as a functional TCSS of M. tuberculosis and elucidated the transcriptional changes in H37Rv and a ΔnarL mutant strain during aerobic growth with or without nitrate. In the presence of nitrate, NarL regulated expression of an ∼30 gene regulon that coincided with DevR-dependent genes (13), indicating possible involvement of DevR in co-regulation with NarL. Mechanistically, we show protein-protein interactions between NarL and DevR RRs in vivo and demonstrate cooperative binding of NarL and DevR to the promoter regions of co-regulated genes like narK2, acg, and Rv3130c. Finally, the data presented here illustrate a novel mode of transcriptional regulation via physical interaction of two DNA-binding RR proteins that has implications regarding integration and fine tuning of signaling pathways in mycobacteria.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
E. coli JM109, HB101, and DH10β strains were used as host strains for genetic manipulations and plasmid constructions. For protein purification, E. coli Origami B strain was used. Wild type M. tuberculosis H37Rv, ΔdosR, and ΔdosSΔdosT mutant strains of H37Rv (henceforth referred to as ΔdevR and ΔdevSΔdosT, respectively) and Mycobacterium smegmatis mc2155 were used in the present study. Detailed plasmid constructions are available upon request to V. M. Table 1 summarizes bacterial strains and plasmids used in this study.
TABLE 1.
List of plasmids and strains used in this study
| Strain/Plasmid | Description | Source |
|---|---|---|
| Strain | ||
| M. tuberculosis H37Rv | ATCC 25618, virulent laboratory strain | ATCC |
| M. smegmatis mc2155 | ept-1 | AECOMa |
| E. coli HB101 | F−, Δ(gpt-proA) 62 leuB6 gln44 ara-14 galK2 lacY1 Δ(mcrC-mrr) rpsL20(Strr) xyl-5 mtl-1 recA13 thi-1 | UCSFb |
| E. coli JM109 | e14− (McrA-940) recA1 endA1 gyrA96 thi-1 941 hsdR17 supE44 relA1 (lac-proAB) | Stratagene |
| E. coli DH10β | F− mcrA Δ(mrr-hsdRMS-mcrBC) Φ80lacZΔM15 ΔlacX74 recA1 endA1 araD139 Δ(araleu) 7697 galUgalKrpsLnupG λ− | Invitrogen |
| E. coli Origami B | F− ompThsdSB(rB− mB−) gal dcm lacY1 aphC gor522::Tn10 trxB (KanR, TetR) | Novagen |
| M. tuberculosis ΔdosR | H37Rv ΔdosR | Ref. 13 |
| M. tuberculosis ΔdosSΔdosT | H37Rv ΔdosSΔdosT | Ref. 40 |
| LIX75 | H37Rv ΔnarL | This study |
| Plasmids | ||
| pPROEx-HTa | N-terminal His6 tag bacterial expression vector | Invitrogen |
| pYUB854 | HygR, cosmid vector used in construction of the phasmid carrying the allelic exchange substrate | Ref. 32 |
| DevScyto | devS gene (from amino acids 378–578) in pProEx-HTc | Ref. 61 |
| DosTcyto | dosT gene (from amino acids 380–573) in pProEx-HTc | Ref. 62 |
| GST-DevR | devR gene in pGEX-4T1 expression vector | Ref. 63 |
| pUAB100 | E. coli-Mycobacterium shuttle plasmid expressing GCN4-mDHFR F[1,2] under hsp60 promoter, Hygr | Ref. 31 |
| pUAB200 | E. coli-Mycobacterium shuttle plasmid expressing GCN4-mDHFR F[3] under hsp60 promoter, Kanr | Ref. 31 |
| pUAB200::devR | devR gene fused with DHFR F[3] fragment | Ref. 31 |
| pUAB200::devS | devS gene fused with DHFR F[3] fragment | Ref. 31 |
| pYA1672 | narL gene in pUAB100 fused with DHFR F[1,2] fragment | This study |
| pYA1673 | narS gene in pUAB200 fused with DHFR F[3] fragment | This study |
| pYA1674 | narL gene in pProEx-HTa expression vector | This study |
| pYA1675 | narS gene (bp 657–1278) in pProEx-HTa expression vector | This study |
| pYA1678 | 5′- and 3′-flanking region of narL gene in pYUB854 | This study |
| pYA1691 | H241Q mutation incorporated in pYA1675 | This study |
| pYA1692 | D61N mutation incorporated in pYA1674 | This study |
| pYA1693 | narL gene fused to mRuby fluorescent protein coding gene at C-terminal end in pProEx-HTa expression vector | This study |
a Dr. William R. Jacobs, Jr. (Albert Einstein College of Medicine, New York, NY).
b Dr. Herbert Boyer (University of California, San Franscisco, CA).
Media, Chemicals, and Culture Conditions
E. coli cultures were grown in 2× YT or Luria-Bertani (LB) broth or on LB-agar plates at 37 °C. M. smegmatis cultures were grown at 37 °C with aeration in Middlebrook 7H9 (Difco) or LB medium supplemented with 0.05% Tween 80 or on LB agar plates. For the mycobacterial protein fragment complementation assay (M-PFC), M. smegmatis strains were plated on Middlebrook 7H11 agar (Difco) supplemented with 0.5% glycerol, 0.5% glucose, and 0.2% Tween 80 and grown at 37 °C. For the nitrate-metabolism model, M. tuberculosis cultures were cultivated in Middlebrook 7H9 medium supplemented with 0.05% Tween 80 and 10% ADS (0.5% albumin, 0.2% dextrose, 0.085% saline) (7H9T medium) with or without 5 mm sodium nitrate (referred to as nitrate) to an A600 of 0.3–0.4 or on Middlebrook 7H10 agar plates at 37 °C. For studies involving a short exposure to nitrate/nitrite (short-pulse model), aerobic cultures of M. tuberculosis were exposed to 5 mm sodium nitrate or 0.5 mm sodium nitrite (referred to as nitrite) for 15 min before processing for RNA isolations. The untreated cultures served as the aerobic controls. For hypoxic growth, M. tuberculosis was subjected to hypoxic growth conditions in sealed standing tubes as described previously (29). Antibiotics and chemicals were added as required at the following concentrations: ampicillin (Amp), 100 μg/ml; kanamycin (Kan), 50 μg/ml; hygromycin (Hyg), 50 and 100 μg/ml for M. smegmatis and M. tuberculosis, respectively, and 150 μg/ml for E. coli; isopropyl β-d-thiogalactopyranoside, 1 mm; and trimethoprim (TRIM), 20 μg/ml. All chemicals were obtained from Sigma-Aldrich unless stated otherwise.
Cloning, Expression, and Purification of NarL and NarS Wild Type and Mutant Proteins
The full-length narL gene (651 bp) encoding a 216-amino acid protein and the cytosolic domain of narS, coding for the catalytic region (657–1278 bp) corresponding to a protein of 206 amino acids (NarScyto) were amplified from M. tuberculosis H37Rv genomic DNA using gene-specific primers containing BamHI/PstI and BamHI/KpnI restriction sites, respectively. Both of the genes were cloned into the pPROEx-HTa expression vector that provides an N-terminal His6 tag for protein purification by affinity chromatography. The H241Q mutation in pYA1675 (NarScyto expression plasmid) and D61N mutation in pYA1674 (NarL expression plasmid) were introduced using the QuikChange site-directed mutagenesis protocol to yield pYA1691 and pYA1692, respectively. The mutants were confirmed by DNA sequencing. The NarL-mRuby-expressing plasmid, pYA1693, was generated in the pPROEx-HTa vector, where full-length NarL was cloned between BamHI and XhoI and mRuby (30) was cloned between XhoI and HindIII, containing the linker region GGATCTGGTGGAGGT between the two open reading frames.
For large scale protein production, each clone was grown at 37 °C, 180 rpm, in 2× YT broth to an A600 of 0.8–0.9 and then induced with isopropyl β-d-thiogalactopyranoside (0.1–1.0 mm). The cultures were incubated for 15–20 h at 12–15 °C. NarL-mRuby, DevScyto, and DosTcyto were expressed and purified as His6-tagged proteins, whereas DevR was purified as a GST-tagged protein. All of the proteins were purified under native conditions by Ni2+-nitrilotriacetic acid or GST affinity chromatography, dialyzed against storage buffer (50 mm Tris-HCl, pH 8.0, 50 mm NaCl, 1 mm DTT, and 50% glycerol), and stored at −20 °C.
Autophosphorylation of Sensor Kinases
150 pmol of purified sensor kinase proteins were incubated in the autophosphorylation buffer (50 mm Tris-HCl, pH 8.0, 50 mm KCl, 10 mm MgCl2) containing 50 μm ATP and 1 μCi of [γ-32P]ATP for the indicated time periods at 30 °C. The reaction was terminated by adding 1× SDS-polyacrylamide gel loading buffer and kept on ice. For analysis, the reaction mixture was resolved on 15% SDS-PAGE at 150 V, and the gel was exposed to an image film (Fujifilm Bas cassette2) followed by phosphorimaging. For quantitative measurements, densitometry was performed using the ImageJ software.
Transphosphorylation of RRs by Sensor Kinases
To study phosphotransfer, 150 pmol of purified sensor kinases were autophosphorylated for 60 min as described above in a reaction volume of 10 μl, and then 150 pmol (or an amount otherwise indicated) of the RR proteins were added to the reaction mixture in a total volume of 20 μl and incubated at 30 °C for the indicated time periods. Reaction was terminated by adding 1× SDS-polyacrylamide gel loading buffer and analyzed as described above.
Surface Plasmon Resonance Analysis
The interaction between the NarL and NarScyto was analyzed using surface plasmon resonance (SPR). Briefly, His6-tagged NarL protein was immobilized on the CM5 chip as per the manufacturer's protocol. Various amounts of His6-tagged NarScyto protein (analyte) were passed over the chip. All the interactions and dilutions were performed in running buffer containing 0.01 m HEPES (pH 8.0), 0.15 m NaCl, 50 μm EDTA, 0.005% (v/v) surfactant P-20 at 25 °C. Each interaction was performed in duplicate and evaluated using a Biacore 3000 instrument (GE Healthcare). BSA was used as an analyte for negative control.
Mycobacterial Protein Fragment Complementation
To investigate possible interactions between NarL and NarS, and NarL and DevRS proteins in vivo, we used the M-PFC system as described previously (31). The plasmid pair, pUAB100 (which expresses Saccharomyces cerevisiae GCN4 fused to domains 1 and 2 of the F portion of mouse dihydrofolate reductase (mDHFR)) and pUAB200 (which expresses GCN4 fused to domain 3 of mDHFR) (hereafter designated as GCN4F[1,2] and GCN4F[3] domains, respectively) homodimerize and serve as the positive control in these experiments. The narL gene was cloned into the episomal mycobacterium-E. coli shuttle plasmid pUAB100 to yield pYA1672, which expresses narL fused to the F[1,2] mDHFR domain. The narS gene was cloned into the integrative mycobacterium-E. coli shuttle plasmid pUAB200 to yield pYA1673, which expresses the NarS protein fused with a F[3] mDHFR domain at its C-terminal end. M. smegmatis was co-transformed with the respective plasmid pairs and plated on 7H11-Kan-Hyg plates to allow growth of all transformants, followed by selection on 7H11-Kan-Hyg-TRIM medium plates to identify positive protein-protein interactions.
Estimation of Nitrate and Nitrite by Flow Injection Analysis
M. tuberculosis H37Rv cultures were aerobically grown in Middlebrook 7H9T medium with or without 5 mm sodium nitrate. After 5 days of growth (A600 of 0.3–0.4), cells were harvested by centrifugation, and the supernatants were filtered before assaying to determine the nitrate (NO3−)/nitrite (NO2−) concentrations. The filtrates were analyzed for the presence of NO3− alone and for total NO3− and NO2− ions using the QuikChem® method 10-107-04-1-C on the QC8000 flow injection analyzer (Lachat Instruments). The amount of NO2− produced by M. tuberculosis cells was obtained by subtracting the NO3− amounts from the total (NO2− + NO3−) estimations. The data are presented as average mg of nitrogen/liter ± S.D. estimations from two independent experiments.
Construction of the M. tuberculosis ΔnarL Mutant Strain
The narL gene (Rv0844c) was deleted in M. tuberculosis H37Rv and replaced with a Hyg resistance cassette by allelic exchange using the specialized transducing mycobacteriophage system (32). Briefly, a recombinant phAE87 phage containing the 5′- and 3′-flanking regions of narL was constructed and transduced in E. coli HB101. The phasmid DNA was electroporated into M. smegmatis mc2155 and plated for mycobacteriophage plaques at 30 °C. A high titer phage lysate prepared from one temperature-sensitive phage plaque was used to infect M. tuberculosis H37Rv as described previously (32). Hygromycin-resistant colonies were selected and further analyzed by PCR and Southern hybridizations for absence of the narL gene and the presence of the hyg gene.
RNA Isolation
Exponentially grown cultures of M. tuberculosis were processed for RNA isolation as described (33). Briefly, cells were harvested by centrifugation, resuspended in 1 ml of TRI reagent (Ambion), and mechanically lysed in a FastPrep FP120 bead beater using 0.1-mm zirconium-silica beads. The cell lysates were processed for RNA extraction according to the manufacturer's specifications. To remove DNA contamination, RNA samples were treated with RNase-free Turbo DNase (Ambion) and verified by PCR analysis of the 16 S rDNA. Total RNA integrity was assessed using RNA 6000 Nano Lab Chip on the 2100 Bioanalyzer (Agilent) according to the manufacturer's protocol. Total RNA purity was assessed by the NanoDrop® ND-1000 UV-visible spectrophotometer (NanoDrop Technologies).
Microarray Hybridization and Quantitative RT-PCR
RNA samples from two independent experiments were analyzed by microarrays as described previously (34). All reagents for microarrays were obtained from Agilent unless specified. Briefly, poly(A) tails were added to the 3′-ends of RNA using the A-plus poly(A) polymerase tailing kit (Epicenter Biotechnologies), and the samples were labeled using the Quick-Amp labeling kit. Each sample (500 ng) was incubated with reverse transcription mix at 42 °C and converted to double-stranded cDNA primed by oligo(dT) with a T7 polymerase promoter. The cDNA was used as template for cRNA generation by in vitro transcription and incorporation of the dye Cy3-CTP and Cy5-CTP. The quality of labeled cRNA was assessed for yields and specific activity, followed by hybridization on a custom M. tuberculosis H37Rv array designed by Genotypic Technology Pvt. Ltd. (AMADID: 023057, Agilent). Cy3- and Cy5-labeled samples were hybridized in Surehyb chambers at 65 °C for 16 h, washed, and scanned using the microarray scanner G2505C. Data extraction from images was done using Feature Extraction software version 10.5.1.1, followed by analysis using the GeneSpring GX version 10 software. Samples were grouped based on the replicates, and genes showing up- or down-regulation >0.6-fold among the samples were identified. Results are reported as the geometric means of log2 expression ratios ± S.D. between the replicates.
Quantitative RT-PCR (qRT-PCR) was performed as described previously (33). Briefly, 200 ng of RNA was reverse-transcribed into cDNA using the iScript cDNA synthesis kit (Bio-Rad). The cDNA was diluted 1:10 and served as the template in real-time PCR using gene-specific primers and SYBR Green dye (Bio-Rad). RNA from two independent experiments was used for expression analysis. For each quantitative PCR experiment, the calculated threshold cycle (CT) was normalized to the CT of the internal 16 S rRNA control (amplified from the same samples) before calculating the -fold change between the test and the control samples. All expression analysis was done using the iQ5 software.
Electrophoretic Mobility Shift Assay
The upstream regions of the narK2, acg, Rv3130c, and Rv1856c genes and the intergenic region between narL and narS open reading frames were PCR-amplified from M. tuberculosis H37Rv genomic DNA template using specific primers. The PCR products were gel-purified using the GeneJET gel extraction kit (Thermo Scientific) and end-labeled with γ-32P using T4 polynucleotide kinase as per the manufacturer's protocol (Thermo Scientific). The fragments were repurified and used directly for binding assays. In an electrophoretic mobility shift assay (EMSA), the labeled DNA was incubated with the indicated amount of proteins for 40 min at 25 °C in the binding buffer containing 25 mm Tris-HCl, pH 8.0, 20 mm KCl, 6 mm MgCl2, 0.10 mg/ml BSA, 0.5% glycerol, 1 mm DTT, 0.5 mm EDTA, 2 mm ATP, and 1 μg of poly(dI-dC). Postincubation, reaction mixtures were separated on a 5% native polyacrylamide gel (pre-run) in 0.5× Tris borate EDTA buffer at 4 °C at 80 V for 2–3 h. The DNA-protein complexes were visualized by phosphorimaging.
For studying the effect of RR phosphorylation on EMSA, DevR and NarL proteins were autophosphorylated for 10 and 45 min, respectively, at 37 °C using 35 mm lithium potassium acetyl phosphate (Sigma) in a reaction buffer containing 50 mm Tris-HCl, pH 8.0, 50 mm KCl, and 10 mm MgCl2 and further incubated with DNA as described above.
Statistical Analyses
Statistical analyses were performed using Student's t test or one-way analysis of variance using GraphPad Prism version 5.0 software. A p value of <0.05 was considered statistically significant.
RESULTS
Genomic Arrangement and Domain Organization of M. tuberculosis Rv0844c-Rv0845 Genes Encoding Putative NarL-NarS TCSS
The M. tuberculosis Rv0844c gene is predicted to encode NarL, a RR belonging to the LuxR/UhpA family of transcriptional regulators (26). Fig. 1A (top) shows a schematic for the location of narL (Rv0844c) and adjacent genes on the M. tuberculosis chromosome. The narL gene is positioned next to Rv0845, which encodes a putative transmembrane sensor kinase (26). Based on the proximity between the two genes, Parish et al. (35) suggested Rv0845 as the cognate kinase for NarL and named it NarS; however, no experimental evidence of signaling between the NarS and NarL proteins has been reported.
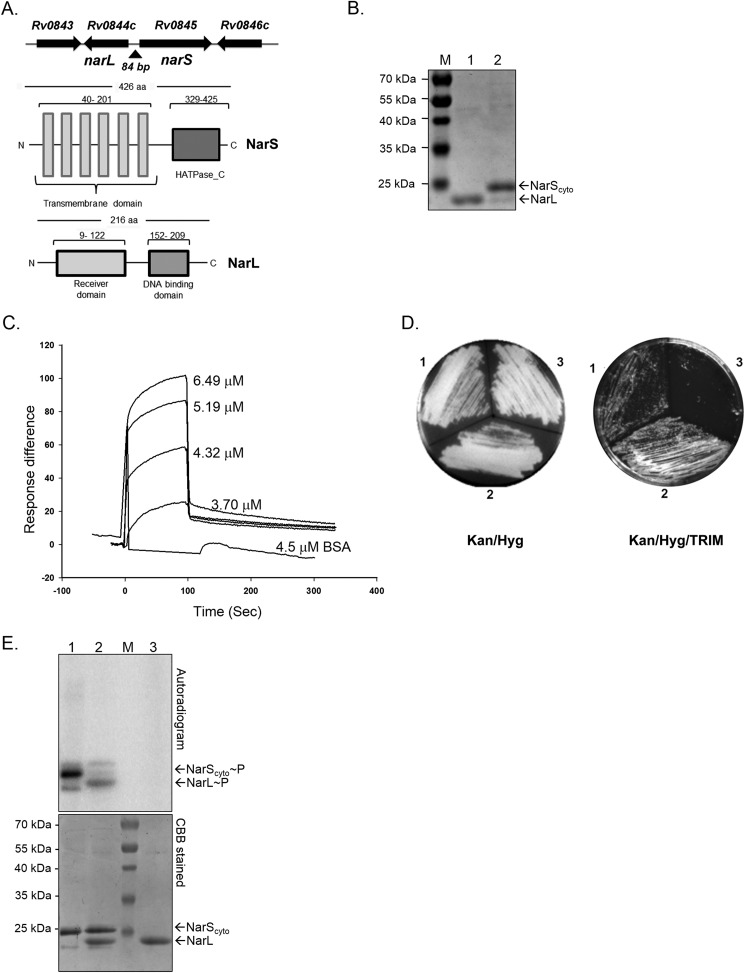
FIGURE 1.
Features and characterization of NarL-NarS two-component regulatory system. A, top, schematic of the genomic location of narL, narS, and adjacent genes on the M. tuberculosis chromosome. Arrows indicate the direction of transcription. Bottom, domain organization of NarL and NarS proteins of M. tuberculosis H37Rv. B, purification and analysis of full-length NarL (lane 1) and cytosolic fragment of NarS (NarScyto) (lane 2) proteins on 15% SDS-PAGE. M, molecular size protein marker C, sensograms of interaction analysis between various concentrations of NarScyto and NarL proteins using SPR. The KD value for NarLS interaction equals 0.765 μm. D, in vivo interactions of NarL and NarS proteins in mycobacteria using the M-PFC system. Left, non-selective Kan/Hyg-containing solid medium allowing growth of all transformants. Right, Kan/Hyg/TRIM-containing solid medium allows growth of positive control (section 1) and NarLF[1,2]/NarSF[3] proteins (section 2) in M. smegmatis. NarLF[1,2]/GCN4F[3](pUAB200) served as a negative control (section 3). E, autophosphorylation of NarScyto and phosphotransfer to NarL. Lane 1, NarScyto autophosphorylated for 60 min; lane 2, NarScyto to NarL phosphotransfer for 5 min; lane 3, NarL incubated with [γ-32P]ATP for 60 min in similar conditions as used for NarScyto alone (negative control). CBB, Coomassie Brilliant Blue.
To identify the organization of conserved domains in the NarS and NarL proteins, their primary amino acid sequences were analyzed using the SMART server (available through the EMBL Web site). The NarS sensor kinase, a protein of 426 amino acids, has six predicted transmembrane domains followed by a 96-amino acid-long histidine kinase-type ATPase catalytic (HATPase_C) domain (Fig. 1A, bottom). Similarly, in the NarL response regulator protein, a DNA-binding helix-turn-helix domain of 57 amino acids in the C-terminal end of the protein was predicted along with a conserved N- terminal receiver domain of 113 amino acids (Fig. 1A, bottom).
NarL and NarS Proteins Constitute a Functional Two-component System
For functional characterization of the TCSS, NarL and NarS proteins were overexpressed and purified from E. coli as His6-tagged proteins. Given the fact that for bacterial HKs, the C-terminal end alone is active in vitro and can be utilized for kinase activity (36) and in view of the six predicted transmembrane domains in the NarS full-length protein (Fig. 1A), we used only the C-terminal catalytic domain of the NarS protein for our studies (henceforth named as NarScyto). Both NarL (predicted Mr 26,000) and NarScyto (predicted Mr 25,000) proteins were obtained in soluble form and confirmed to be >90% pure by SDS-PAGE analysis (Fig. 1B).
To determine whether these proteins interact with each other, we performed in vitro SPR analysis using NarL protein immobilized on CM5 chip, and the NarScyto protein or BSA was used as analyte. As is evident from Fig. 1C, when NarS protein was used, there was an increase in the SPR indicated by response units or difference on the y axis, unlike BSA, which did not interact with the NarL protein. We observed a concomitant increase in the response units as the concentration of NarS was increased, suggesting interaction of these proteins in the basal condition (Fig. 1C). We also determined the dissociation constant (KD) for NarL-NarS interaction. A KD value of 0.765 μm was recorded. The kon (ka) value was determined to be 3.69 × 103 m−1 s−1, and the koff (kd) was 2.82 × 10−3 s−1, indicating rapid dissociation of the two proteins. Such values are typical for other mycobacterial TCSS proteins (37, 38), indicating that although the interaction is very transient, it is of very high affinity.
Given the above results, we investigated the interaction between NarL and NarS proteins within mycobacterial cells using the M-PFC assay. Positive interactions between the two proteins of interest, independently fused with mDHFR domains F[1,2] or F[3], were scored by the functional reconstitution of mDHFR activity and the resulting TRIM resistance of the transformants. As shown in Fig. 1D, the following combinations were evaluated: GCN4F[1,2] (pUAB100)/GCN4F[3] (pUAB200) (section 1, positive control), NarLF[1,2] (pUAB100::narL)/NarSF[3] (pUAB200::narS) (section 2), and NarLF[1,2] (pUAB100::narL)/GCN4F[3] (pUAB200) (section 3, negative control). All co-transformants grew on agar plates without TRIM (Fig. 1D, left). On TRIM-containing agar plates, interactions between the GCN4F[1,2] and GCN4F[3] protein domains in pUAB100/pUAB200 vectors, respectively (positive control), allowed growth of M. smegmatis (Fig. 1D, section 1), whereas no growth was observed with negative control (Fig. 1D, section 3). M. smegmatis co-transformed with the NarLF[1,2]/NarSF[3] plasmid pair was also able to grow on TRIM-selective medium (Fig. 1D, section 2), confirming direct intracellular interactions between NarL and NarS proteins in mycobacteria.
To establish signal transduction between the NarS HK and NarL RR proteins, we performed in vitro phosphorylation assays. Activity analysis of purified NarScyto protein was performed in a typical sensor kinase autophosphorylation reaction (29). We found that the NarScyto protein is capable of autocatalyzing the transfer of 32P-labeled γ-phosphate from ATP onto itself. The occurrence of phosphorylation is detected by the presence of radiolabeled protein band on the autoradiogram (Fig. 1E, lane 1, top). The assay confirmed that the Rv0845/narS gene encodes a functional sensor kinase protein. To establish signaling with NarL RR, a phosphotransfer assay was performed. The assay involved direct transfer of a phosphoryl moiety from the active kinase to a conserved residue on the RR protein (39). To do this, 150 pmol of NarScyto was autophosphorylated for 60 min, before NarL was added to the reaction. Equimolar amounts of purified NarL and [γ-32P]NarS were incubated together to facilitate phosphotransfer. The transfer of γ-32P-labeled phosphoryl moiety from NarS to NarL is detected by the appearance of a labeled band corresponding to the position of NarL and also a decrease in signal in the NarScyto protein in the autoradiogram (Fig. 1E, lane 2). We observed robust phosphorylation of the NarL protein, confirming that NarL serves as a cognate RR for NarS. No signal was detected on NarL in the absence of NarS, validating that the reaction occurred specifically due to the phosphorylation via the sensor kinase and not directly by ATP (Fig. 1E, lane 3). After autoradiography, the gel was stained with Coomassie Brilliant Blue to visualize the position of the radiolabeled proteins (Fig. 1E, bottom). Taken together, the direct intracellular interaction between NarL and NarS proteins in vivo in the M-PFC assay and in vitro by SPR and NarS-mediated phosphorylation of NarL confirmed that both Nar proteins (i.e. Rv0844c/Rv0845) are active and constitute a functionally competent two-component signal transduction system in M. tuberculosis.
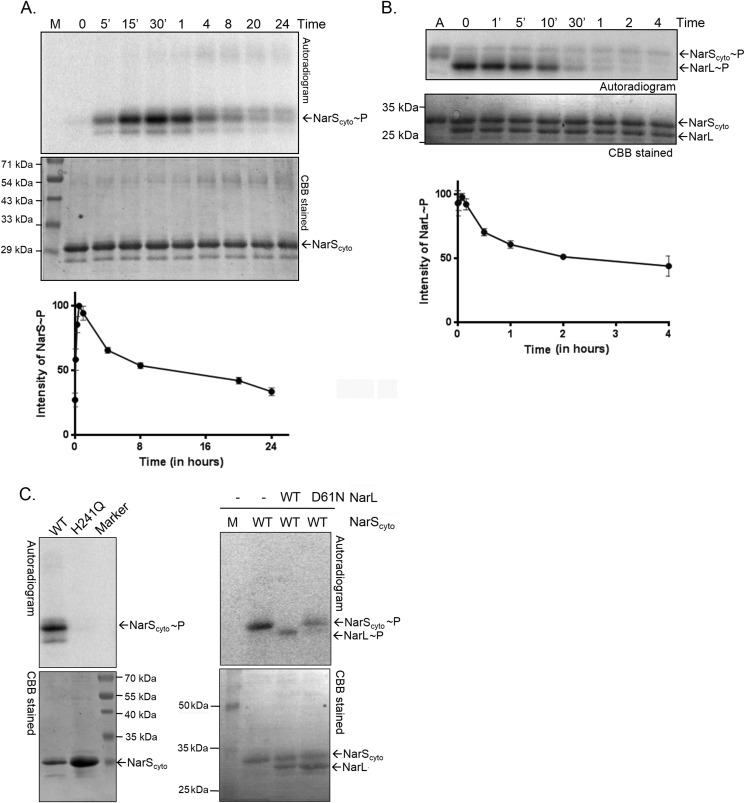
Phosphorylation Kinetics of NarS and NarL Proteins
Autophosphorylation of NarScyto and phosphotransfer to NarL was followed over a time course experiment. Like most HKs, NarS underwent rapid autophosphorylation (Fig. 2A, top), and maximal incorporation of the γ-32P phosphoryl moiety occurred within 30–60 min, after which a progressive decrease in signal was observed (Fig. 2A). In agreement with typical phosphotransfer reactions (39), we observed phosphorylation of the NarL protein almost instantaneously (Fig. 2B). Sequence comparison of the Nar proteins with other HKs and RR proteins allowed us to identify His241 and Asp61 as the putative phosphorylation sites in NarS and NarL proteins, respectively. To confirm, H241Q substitution in NarScyto and D61N in NarL proteins were introduced by site-directed mutagenesis, and the mutant proteins were tested for their phosphorylation potential as described above. As anticipated, the mutated forms of both the proteins were defective in their phosphorylation capacities (Fig. 2C). These observations confirm that the γ-32P from labeled ATP is transferred to the conserved His241 residue of NarS during autophosphorylation, and the phosphotransfer reaction involves direct transfer of the phosphoryl moiety to the conserved Asp61 residue on the NarL RR protein.
FIGURE 2.
Phosphorylation kinetics of the NarS and NarL proteins. Auto- and transphosphorylation reactions were set up as described under “Experimental Procedures.” Top, autoradiograms; the corresponding Coomassie Brilliant Blue (CBB)-stained gel is located below each autoradiogram. A, NarScyto autophosphorylation time course. B, time course of phosphotransfer of phosphoryl group from NarScyto∼P to NarL. M, molecular size protein marker; A, autophosphorylation control. The 0 min time point is technically ∼10 s due to experimental limitations. Bottom panel, densitometric plot of phosphorylated protein from the autoradiogram as a function of time. C, characterization of H241Q mutant of NarS HK (left) and D61N mutant of NarL (right) in phosphorylation reactions as described for Fig. 1E. Wild type (WT) NarL or NarScyto served as control in the assays.
Aerobically Grown M. tuberculosis Metabolizes Nitrate to Secrete Nitrite into Extracellular Medium
To study the role of the NarL/NarS TCSS in the regulation of nitrate metabolism in mycobacteria, we established experimental conditions that were marked by metabolism of nitrate to produce nitrite during aerobic growth (nitrate-metabolism model). In this model, M. tuberculosis H37Rv was grown aerobically in Middlebrook 7H9T medium supplemented with 5 mm sodium nitrate (equal to 70 mg of nitrogen/liter). Logarithmic phase cultures of H37Rv grown with or without nitrate were analyzed for 1) utilization of nitrate or decrease in nitrate concentrations in the medium over time and 2) secretion of nitrite into the medium by flow injection analysis. Under these experimental conditions, ∼12.5% of the nitrate from the medium is metabolized by H37Rv after 5 days of growth (Table 2). Compared with the controls grown without added nitrate, M. tuberculosis H37Rv cultivated in the presence of 5 mm nitrate showed an ∼293-fold increase in the production of nitrite (Table 2). The medium alone with 5 mm sodium nitrate served as the control for spontaneous generation of nitrite from nitrate. We confirmed that in the absence of any bacteria, there was no nitrite in the culture medium containing 5 mm nitrate, indicating that nitrite production occurred only as a result of nitrate reduction by M. tuberculosis.
TABLE 2.
Estimation of nitrate and nitrite concentrations in medium by flow injection analysis
| Ion | Mediuma + nitrate | M. tuberculosis H37Rv − nitrate | M. tuberculosis H37Rv + nitrate |
|---|---|---|---|
| Nitrate (NO3−) (mg of nitrogen/liter)b | 68.6 ± 2.54 | 1.416 ± 0.08 | 60.02 ± 1.5 |
| Nitrite (NO2−) (mg of nitrogen/liter)b | NDc | 0.024 ± 0.002 | 7.04 + 0.16 |
a Middlebrook 7H9T medium + 5 mm nitrate (∼70 mg of nitrogen/liter).
b Data are presented as average values (mg of nitrogen/liter) ± S.D. of two independent estimations after 5 days of M. tuberculosis growth (A600 ∼0.3–0.4).
c ND, not detected.
NarL Regulates Gene Expression during Aerobic Nitrate Metabolism
To investigate the regulatory role of NarL, we constructed an M. tuberculosis ΔnarL mutant strain, designated LIX75, with a hygromycin resistance gene (hyg) replacing narL. The presence of the hyg gene in LIX75 was confirmed by Southern hybridization using a hyg-specific probe (data not shown). Initial phenotypic analysis of the ΔnarL mutant did not reveal any defects in terms of in vitro growth in broth cultures with or without 5 mm nitrate or the ability to metabolize exogenous nitrate to nitrite during aerobic growth (NO2−, 6.8 ± 0.11 mg of nitrogen/liter for ΔnarL versus 7.04 ± 0.16 mg of nitrogen/liter for H37Rv).
We performed transcriptional profiling of H37Rv and ΔnarL mutant strains during aerobic growth in the presence or absence of 5 mm nitrate by microarray analysis. A number of genes were differentially regulated in the ΔnarL mutant grown in the presence of nitrate. This suggested that NarL has a distinct regulatory role in nitrate metabolism (n = 45; Up = 15, Down = 30) but not during aerobic growth without nitrate (n = 5; Up = 1, Down = 4), where n is the number of differentially regulated genes. The complete normalized microarray data from two independent replicates are provided in supplemental data sets S1 and S2. The genes that were down-regulated in the ΔnarL mutant under the two experimental conditions are separately listed in Table 3. Essentially, in aerobically grown cultures with nitrate, ∼30 genes were found to be down-regulated in the narL mutant, indicating that these genes require NarL for their expression during nitrate metabolism (Table 3).
TABLE 3.
Genes down-regulated in ΔnarL mutant relative to H37Rv during aerobic growth (with or without nitrate)
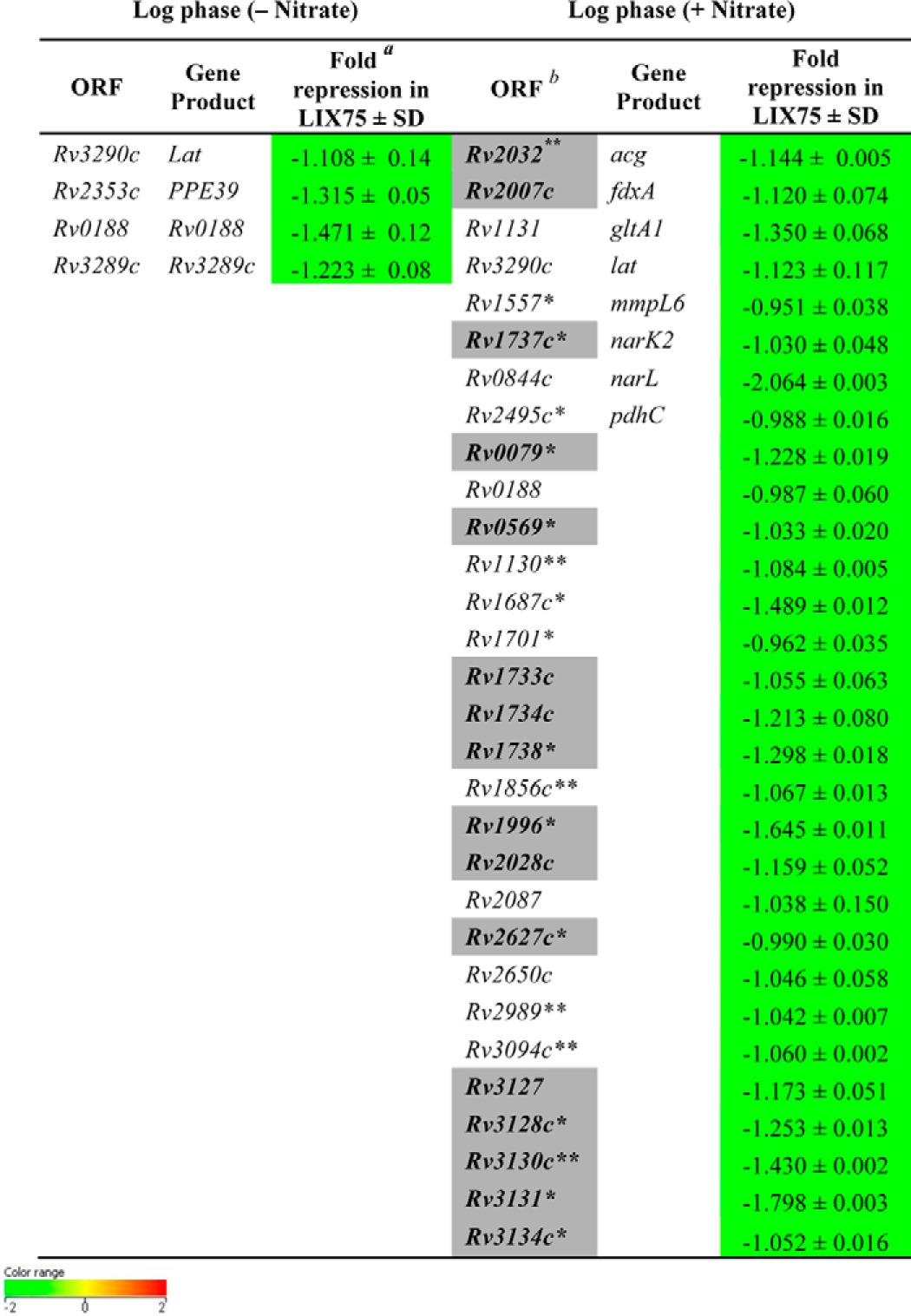
a Geometric mean of log 2 ratio of expression in ΔnarL mutant relative to H37Rv under a specific condition ± S.D.
b Shaded boxes represent overlap with the hypoxia-induced and NO-induced DevR regulon (shown in boldface type) (8, 13). Green boxes indicate -fold change <−0.6 value in ΔnarL mutant relative to H37Rv.
*(p < 0.05), ** (p < 0.01), and *** (p < 0.001) (Student's t test) represent significant difference between ΔnarL mutant and H37Rv.
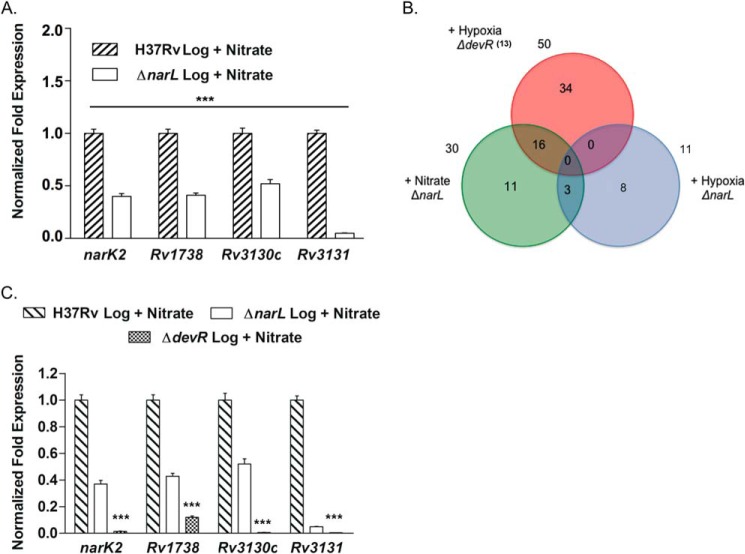
For four genes that are down-regulated in the ΔnarL mutant (narK2, Rv1738, Rv3130c, and Rv3131), we validated the microarray results by qRT-PCR (Fig. 3A). Comparisons between H37Rv and ΔnarL mutant grown in the presence of nitrate revealed 2-fold down-regulation of the three genes and ∼20-fold down-regulation for Rv3131 gene in the mutant (Fig. 3A). These results validate the microarray findings and support the role of NarL as a regulator of gene expression during aerobic growth in the presence of nitrate.
FIGURE 3.
Overlap between NarL-regulon and hypoxia-induced DevR regulon. A, qRT-PCR expression analysis of narK2, Rv1738, Rv3130c, and Rv3131 in RNA isolated from wild type H37Rv and ΔnarL mutant strain cultivated in the presence of nitrate. B, Venn diagram representation of the number of genes that are down-regulated in the ΔnarL mutant and require NarL for expression during aerobic nitrate metabolism (green circle) and 48 h hypoxic (blue circle) growth conditions. An overlap of NarL-dependent genes with the hypoxia-inducible DevR-dependent genes (13) is shown (red circle). C, qRT-PCR analysis of narK2, Rv1738, Rv3130c, and Rv3131 genes in wild type H37Rv, ΔnarL, and ΔdevR mutant strains in the presence of nitrate. Expression is normalized using the 16 S rRNA gene as an internal control. The -fold change in gene expression in the ΔnarL mutant relative to H37Rv grown under a specific growth condition is presented as the mean ± S.D. (error bars) of data from two independent experiments. The baseline expression of the target genes in H37Rv under the specified growth condition is set to 1.0. ***, p < 0.001.
A Subset of NarL-regulated Genes Are Co-regulated by the DevR Response Regulator
Further analysis of the genes that are regulated by NarL revealed a striking overlap with the DevR regulon known to be induced by hypoxia (13) and NO (8). As shown by a Venn diagram, of 30 NarL-dependent genes, 16 belong to the DevR regulon as well (Fig. 3B and Table 3, shaded boxes), suggesting that both NarL and DevR may co-regulate the expression of these genes. This led us to compare expression profiles of ΔnarL mutant and H37Rv under hypoxic conditions (supplemental data set S3). Interestingly, no overlap of the DevR regulon with the NarL-regulated genes under hypoxia was observed (Fig. 3B), suggesting that co-regulation occurred only during aerobic nitrate metabolism. We confirmed our hypothesis by comparing the expression of narK2, Rv1738, Rv3130c, and Rv3131 in a ΔdevR mutant strain aerobically grown in the presence of nitrate with similarly grown ΔnarL mutant strain. Higher down-regulation of narK2, Rv1738, Rv3130c, and Rv3131 gene expression was observed in the absence of DevR (Fig. 3C). These results explained the partial down-regulation of these genes in the ΔnarL mutant strain where wild type DevR is present. Together, these observations indicate co-regulatory functions of NarL and DevR RRs in regulating expression of a subset of NarL regulon genes during aerobic nitrate metabolism.
DevRS Two-component System and Nitrate Metabolism
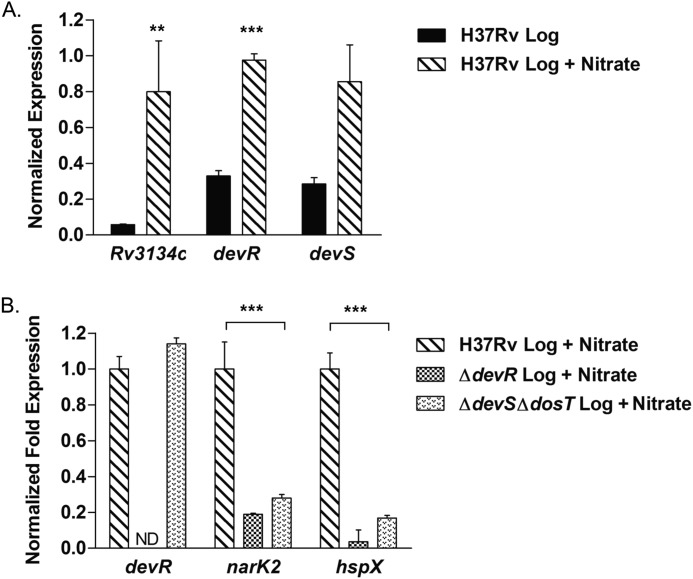
In light of the microarray results, we compared expression of the Rv3134c-devR-devS operon in M. tuberculosis aerobic cultures with or without 5 mm nitrate by qRT-PCR. Our results indicate that the Rv3134c-devR-devS operon is indeed induced in the presence of nitrate during aerobic growth, with the -fold induction being most significant for Rv3134c (13.8-fold) followed by devR (3.0-fold) and devS (3.0-fold) in comparison with the control without nitrate (Fig. 4A).
FIGURE 4.
Rv3134c-devR-devS operon is activated during aerobic nitrate metabolism. A, expression of Rv3134c-devR-devS genes was determined in M. tuberculosis H37Rv aerobic cultures grown in 7H9T medium with or without nitrate by qRT-PCR. M. tuberculosis H37Rv grown in the absence of nitrate served as a control. B, induction of Rv3134c-devR-devS operon under nitrate metabolism conditions is phosphorylation-dependent. The transcript levels of devR and two well established DevR-regulated genes, narK2 and hspX, were analyzed by qRT-PCR in M. tuberculosis H37Rv, ΔdevR, and ΔdevSΔdosT cultures cultivated in the presence of nitrate. The expression was normalized using 16 S rRNA gene, and the -fold change in expression of all genes was calculated as described for Fig. 3. ** and ***, p < 0.01 and p < 0.001, respectively. Error bars, S.D.
We further investigated whether the induction of the Rv3134c-devR-devS operon and DevR-regulated genes during aerobic nitrate metabolism was dependent on the activation of DevR protein. Previously, it has been shown that in an M. tuberculosis ΔdevSΔdosT mutant strain, DevR is not phosphorylated, and all DevR-mediated induction of target genes is abrogated (40, 41). To examine whether activation of DevR through DevS and/or DosT kinases is needed under our experimental conditions, we analyzed the transcript levels of key DevR-regulated genes: devR, narK2, and hspX in wild type H37Rv and ΔdevR and ΔdevSΔdosT mutant strains. Compared with H37Rv, narK2 and hspX expression was significantly reduced in the ΔdevR strain (Fig. 4B). Most importantly, despite the presence of devR-specific transcripts in the ΔdevSΔdosT mutant strain, no up-regulation of narK2 and hspX genes was observed (Fig. 4B), indicating a requirement for DevR activation by DevS/DosT kinases for regulation of target gene expression during aerobic nitrate metabolism. In addition, these observations also rule out the possibility of cross-activation of DevR by other HKs, particularly NarS.
Nitrate Versus Nitrite as a Metabolic Signal
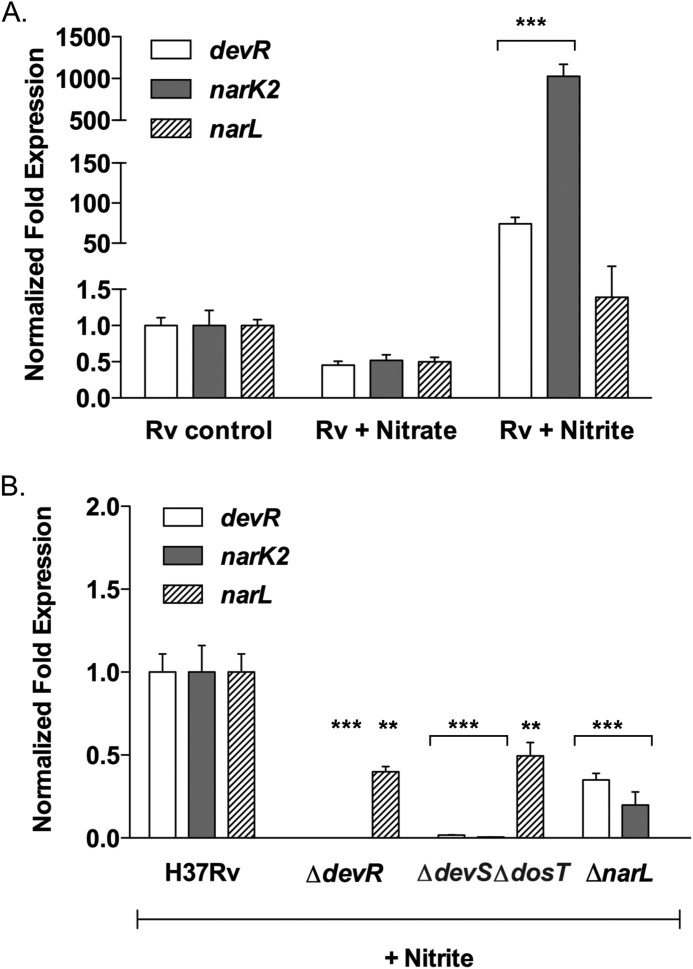
Given our observations that M. tuberculosis grown in the presence of nitrate metabolizes it by reducing it to nitrite and that DevR and NarL co-regulate a transcriptional response in these conditions, the activating signal could either be nitrate or nitrite. To investigate the role of nitrate and nitrite as signals, we analyzed the transcription changes in narL, devR, and a shared target gene, such as narK2, in M. tuberculosis cultures after a short exposure (15 min) to 5 mm nitrate or 0.5 mm nitrite (short-pulse model). This is different from the nitrate metabolism model, where the bacilli are grown in medium supplemented with nitrate for 5 days. As seen in Fig. 5A, short exposure to nitrate resulted in repression of narL, devR, and narK2 genes in H37Rv. Interestingly, significant induction of devR (∼74.0-fold) and narK2 (∼1026.0-fold) expression in H37Rv exposed to nitrite was observed, whereas narL was only moderately induced (∼1.4-fold) (Fig. 5A). These findings implicate nitrite as a metabolic cue and confirm that it is not the presence of nitrate in the medium per se but the production of nitrite by mycobacteria that enables induction of DevR- and NarL-regulated genes in the nitrate metabolism model.
FIGURE 5.
Nitrite, a metabolic signal. The expression of devR, narK2, and narL was determined in logarithmic phase cultures of M. tuberculosis H37Rv after a short exposure to 5 mm nitrate or 0.5 mm nitrite (short-pulse model) (A) and in M. tuberculosis H37Rv, ΔdevR, and ΔdevSΔdosT cultures treated with 0.5 mm nitrite for 15 min (B). Analysis was done as described for Fig. 3. Untreated M. tuberculosis H37Rv served as a control. ** and ***, p < 0.01 and p < 0.001, respectively. Error bars, S.D.
In view of the possibility that physiologically derived nitrite could be the activator, we analyzed the expression of devR, narL, and narK2 genes in H37Rv, ΔdevR, ΔdevSΔdosT, and ΔnarL strains by qRT-PCR after short exposure to nitrite. Unlike wild type H37Rv (Fig. 5A), no significant induction of devR, narK2, and narL genes in the ΔnarL, ΔdevR, and ΔdevSΔdosT mutant strains was observed (Fig. 5B). We were able to detect a modest level of narL expression in the ΔdevR and ΔdevSΔdosT mutant strains; however, it was still significantly reduced compared to H37Rv exposed to nitrite (Fig. 5B). Interestingly, despite some narL expression, devR and narK2 induction is completely abolished in ΔdevR and ΔdevSΔdosT strains, suggesting a primary role of DevR activation in the regulation of gene expression upon nitrite exposure. This allowed us to conclude that the increased expression of narK2 and devR observed in H37Rv as a result of exposure to nitrite is due to activation of the DevRS/DosT system. We noted that the presence of a fully functional DevRS/DosT system in a ΔnarL mutant did not restore devR and narK2 expression to wild type levels (Fig. 5B), indicating that both DevR and NarL RRs are required for the full transcriptional response to nitrite, although NarL involvement appears to be secondary.
Cross-phosphorylation between DevS/DosT/DevR and NarS/NarL Two-component Regulatory Systems
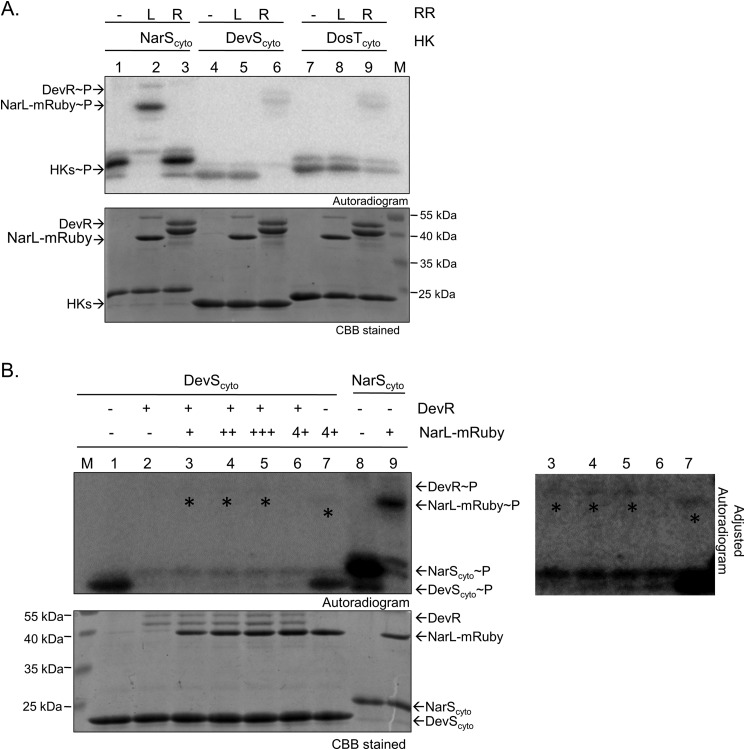
Because M. tuberculosis DevR RR is known to interact with cognate as well as non-cognate HKs (40), it is imperative from the perspective of this study to investigate the possibility of cross-phosphorylation of DevR and NarL RRs by DevS, DosT, and/or NarS sensor kinases. We performed in vitro phosphotransfer experiments with phosphorylated NarScyto, DevScyto, and DosTcyto with DevR or NarL proteins. Because NarL is of similar size as DevScyto and DosTcyto, the separation on SDS-PAGE was suboptimal, and findings were not conclusive (data not shown). Thus, to facilitate clear resolution of proteins, we generated a tagged variant of NarL with mRuby fluorescent protein fused to its C-terminal end (Mr ∼52600 Da) and tested its phosphorylation through DevS/DosT. No cross-phosphorylation was identified between DevS/DosT and NarL-mRuby (Fig. 6A, lanes 5 and 8) or between NarS and DevR (Fig. 6A, lane 3) in a 5-min time window. For all three kinases, their cognate interactions with their specific RRs served as positive controls for the assays (Fig. 6A, lanes 2, 6, and 9). In contrast, when phosphorylated DevScyto was used as a phosphodonor, we could detect weak phosphorylation of NarL-mRuby in 20 min (Fig. 6B, lane 7 and the long exposure image), similar to what has been reported recently (42). To establish whether cognate interaction between DevR-DevS is preferred over non-cognate NarL-DevS interaction, we performed competition experiments wherein the potential of phosphotransfer from DevS to NarL was tested in the presence of a limited amount of DevR and increasing amounts of NarL protein. In none of the tested conditions was phosphorylation of NarL-mRuby detectable in the presence of DevR (Fig. 6B, lanes 3–5 and the long exposure image), suggesting that the weak phosphotransfer from DevScyto to NarL-mRuby (Fig. 6B, lane 7) may not be physiologically relevant.
FIGURE 6.
Analysis of phosphorylation-mediated cross-talk between DevS/DosT/DevR and NarS/NarL two-component systems. A, cross-phosphorylation analysis through DevS, DosT, and NarS sensor kinases. DevScyto∼P, DosTcyto∼P, or NarScyto∼P (150 pmol) was incubated with an equimolar amount of DevR or NarL-mRuby for 5 min to enable phosphotransfer as indicated. L, NarL; R, DevR; M, marker. B, phosphotransfer competition experiments from DevScyto∼P to DevR and NarL-mRuby. DevScyto∼P (150 pmol) was incubated with various amounts of DevR or NarL-mRuby or both for 20 min. Lane 1, DevScyto∼P alone; lanes 2–6, phosphotransfer from DevScyto∼P to 0, 75, 100, 125, and 150 pmol of NarL-mRuby, respectively, in the presence of 15 pmol of DevR; lane 7, phosphotransfer from DevScyto∼P (150 pmol) to NarL-mRuby (150 pmol) in the absence of DevR; lane 8, autophosphorylation of NarScyto; lane 9, phosphotransfer from NarScyto∼P to NarL-mRuby (75 pmol) for 5 min. An overexposed image is shown in the lower panel marking the presence of phosphorylated RRs that are not evident in short exposure shown on the left.
NarL as a DNA Binding Transcriptional Regulator
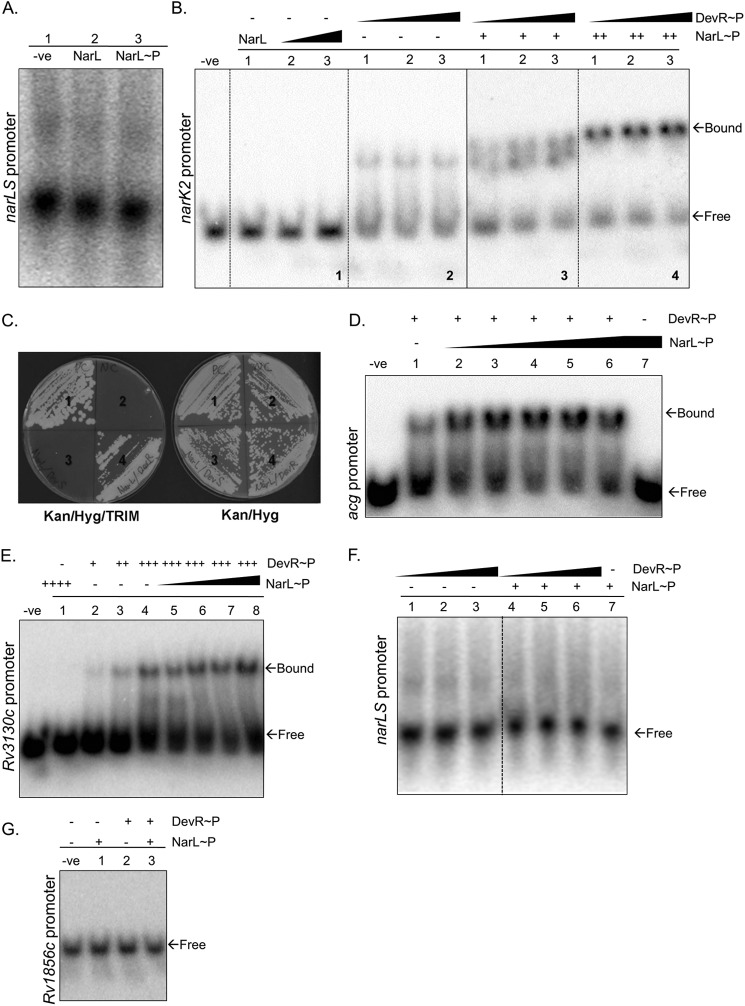
Most M. tuberculosis TCSSs are autoregulated by their respective RR proteins (5); thus, we first tested the ability of NarL to autoregulate narL and narS genes. As shown in Fig. 1A, narL and narS genes are divergently transcribed and have an 84-bp intergenic region (Fig. 1A). We tested the binding of phosphorylated and non-phosphorylated NarL with the 102-bp intergenic region (from −90 to +11 bp from the narS gene) by EMSA. We failed to detect any DNA-protein complex under the conditions tested (Fig. 7A), indicating that this system is perhaps not autoregulated or may require some other regulator/co-factor. Because the narK2 gene is down-regulated in the ΔnarL mutant (Table 3), it was anticipated that NarL may bind to the narK2 promoter region to regulate its expression. In contrast to this expectation, neither NarL (Fig. 7B, panel 1, lane 1) nor NarL∼32P showed any binding to the promoter of the narK2 gene (Fig. 7B, panel 1, lanes 2 and 3). Interestingly, we obtained similar results with three other genes (acg, Rv1856c, and Rv3130c) of the NarL regulon (Fig. 7, D, E, and G), two of which also belong to the DevR regulon. Thus, we concluded that NarL by itself does not bind DNA to regulate expression of these genes.
FIGURE 7.
Cooperative interaction between NarL-DevR RRs. A, EMSA of the intergenic promoter region of narL and narS genes (102 bp; −90 to +11; 1.2 pmol) with NarL protein. Lane 1, probe alone; lane 2, NarL (9 μg); lane 3, NarL∼P (9 μg). B, EMSA showing cooperative effect of NarL and DevR on 4.3 pmol of narK2 promoter region (326 bp; −305 to +20). Lane −ve, probe alone; panel 1, lane 1, 15 μg of NarL; lanes 2 and 3, increasing concentration of NarL∼P (30 and 45 μg, respectively); panel 2, lanes 1–3, increasing concentration of DevR∼P (49, 72, and 100 ng, respectively); panel 3, lanes 1–3, increasing concentration of DevR∼P (49, 72 and 100 ng respectively) with 15 μg of NarL∼P (denoted as +); panel 4, lanes 1–3, increasing concentration of DevR∼P (49, 72, and 100 ng, respectively) with 30 μg of NarL∼P (denoted as ++). C, M-PFC assay showing interaction between the NarL and DevR RR proteins on Kan/Hyg/TRIM and Kan/Hyg agar plates. Shown are positive control (section 1), negative control (section 2), NarLF[1,2] and DevSF[3] (section 3), and NarLF[1,2] and DevRF[3] (section 4). D, EMSA showing the cooperative effect of NarL and DevR on the Rv2032 (acg) promoter region (210 bp; −199 to +11; 0.7 pmol). Lane −ve, probe alone; lane 1, DevR∼P (28 ng); lanes 2–6, increasing concentration of NarL∼P (0.75, 1.5, 2.25, 3.0, and 3.75 μg) with 28 ng of DevR∼P; lane 7, NarL∼P (3.75 μg). E, EMSA showing cooperative effect of NarL and DevR on the Rv3130c promoter region (188 bp; −185 to +3; 0.4 pmol). Lane −ve, probe alone; lane 1, 3 μg of NarL∼P (denoted as ++++); lanes 2–4, DevR∼P (7, 14, and 28 ng, denoted as +, ++, and +++, respectively); lanes 5–8, increasing concentration of NarL∼P (0.75, 1.5, 2.25, and 3.0 μg) with 28 ng of DevR∼P. F, EMSA showing the effect of NarL and DevR on the intergenic promoter region of narL and narS (102 bp; −90 to +11; 1.2 pmol). Lanes 1–3, DevR∼P (20, 40, and 56 ng, respectively); lanes 4–6, DevR∼P (20, 40, and 56 ng, respectively) with 27 μg of NarL∼P (denoted as +); lane 7, 27 μg of NarL∼P. G, EMSA showing the effect of NarL and DevR on the Rv1856c promoter region (350 bp; −350 to −1; 0.2 pmol). Lane −ve, probe alone; lane 1, 3.75 μg of NarL; lane 2, 28 ng of DevR∼P; lane 3, 3.75 μg of NarL and 28 ng of DevR∼P.
NarL Interacts with DevR in Vivo and Cooperative Interactions Enable Co-regulation of Gene Expression
In view of the inability of NarL/NarL∼P to bind to the promoter regions of sentinel NarL regulon genes, we investigated whether this co-regulation is mediated by DevR-NarL interaction and/or DevR-NarL-DNA interaction. To test both of these possibilities, DNA-protein and protein-protein interactions between DevR and NarL and with target gene promoter regions were analyzed.
The M-PFC assay was used to test for interactions between NarL and DevR in vivo. For these experiments, either of the pUAB200::devR and pUAB200::devS plasmids, which encode DevR and DevS proteins, respectively, fused with a F[3] domain at their C-terminal ends (31), were co-transformed with pUAB100::narL into M. smegmatis. Plasmid pairs GCN4F[1,2](pUAB100)/GCN4F[3](pUAB200) and pUAB100::narL/GCN4F[3](pUAB200) served as positive and negative control, respectively. As shown in Fig. 7C, robust growth of all transformants was observed on Kan/Hyg plates. The M. smegmatis co-expressing NarL[F1,2]/DevR[F3] (Fig. 7C, section 4) and the positive control (section 1) grew successfully on TRIM-containing plates, whereas the negative control (section 2) along with M. smegmatis strain with NarLF[1,2]/DevSF[3] (section 3) failed to grow on TRIM plates. Given the faint phosphotransfer signal observed earlier between DevS and NarL (Fig. 6B), the absence of DevS-NarL interaction in the M-PFC assay is not surprising. These results strongly suggest a positive interaction between the NarL and DevR RRs within mycobacteria, which may be a major factor directing the co-regulatory effects of NarL and DevR RRs.
To understand the underlying mechanism of NarL- and DevR-mediated co-regulation, we performed EMSA with promoter regions of three genes that were down-regulated in ΔnarL strain (Table 3) and are also a part of the DevR regulon (Table 3, shaded regions). We selected narK2, Rv3130c, and acg genes because DevR binding to the promoter regions of these genes has already been established (41, 43, 44). As described above, NarL or NarL∼P did not cause a mobility shift at any concentration (Fig. 7, B (panel 1), D (lane 7), and E (lane 1)), but low concentrations of DevR showed marginal shifts for all promoter regions tested (Fig. 7, B (panel 2), D (lane 1), and E (lane 2)). Interestingly, when both NarL∼P and low concentrations of DevR∼P protein were tested together on these promoters, enhanced mobility retardation was detected when compared with lanes where the individual proteins were tested. Compare panels 1 and 2 in Fig. 7B with panels 3 and 4; lanes 1 and 7 in Fig. 7D with lanes 2–6; and lanes 1–4 in Fig. 7E with lanes 5–8. The signal intensity for the protein-DNA complex increased concomitantly with increasing concentrations of NarL (Fig. 7, B, D, and E). These results suggest that the two proteins can co-regulate gene expression by cooperative binding at the same promoter.
In light of these observations, we revisited EMSAs of the narLS intergenic region using both NarL∼P and DevR∼P proteins (Fig. 7F). We still failed to detect any binding at the narLS promoter region. As another control, we analyzed the effect of cooperative binding on the promoter of Rv1856c, which belongs only to the NarL regulon and not the DevR regulon. Here also we did not observe any shift by co-incubation with DevR and NarL (Fig. 7G), confirming that the co-regulatory effect is not nonspecific; rather it is restricted to genes that are common between the NarL- and DevR regulons.
Co-regulation Is Independent of NarL Phosphorylation but Requires DevR Activation
Next, we tested whether this phenomenon was affected by the phosphorylation status of NarL. Thus, EMSAs with the acg (Fig. 8A) and narK2 (Fig. 8B) promoters were performed using DevR along with wild type NarL or phosphorylation-defective NarLD61N mutant protein. As shown in Fig. 8A, the effect of phosphorylation-defective NarLD61N was similar to the wild type NarL protein (Fig. 8A, lanes 3 and 4). Similarly, there was no shift observed with the narK2 promoter in the presence of NarLD61N with DevR (Fig. 8B, lane 2); however, we observed mobility retardation in the presence of NarLD61N and DevR∼P (Fig. 8B, lanes 3 and 4). The data suggest that NarL phosphorylation is not essential for co-regulating gene expression with DevR∼P. Collectively, these observations prove three things: first, that NarL and DevR RRs physically interact in vivo; second, that cooperative interactions enable co-regulation of the common target genes; and third, that DevR phosphorylation is an essential requirement.
FIGURE 8.
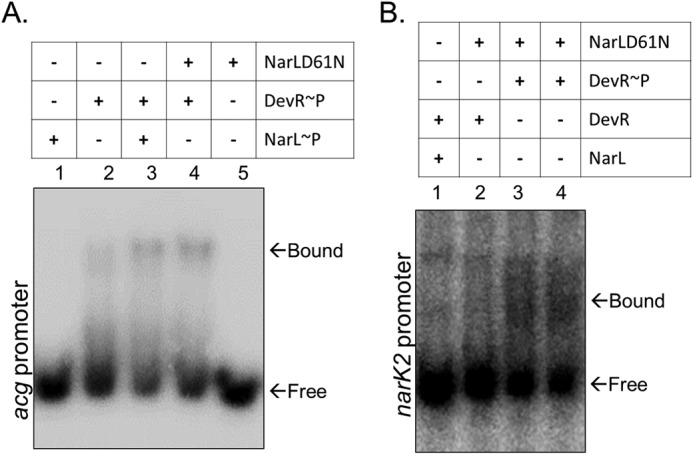
Co-regulation is independent of NarL phosphorylation but requires DevR activation. A, role of NarL phosphorylation in cooperative binding of NarL and DevR on the acg (Rv2032) promoter region. NarL or NarLD61N (1.5 μg) and DevR (28 ng) proteins were used in the EMSAs as depicted. B, role of NarL phosphorylation in cooperative binding of NarL and DevR on the narK2 promoter. All lanes contain 2.0 pmol of narK2 probe. NarL or NarLD61N (9 μg) and DevR (40 ng) proteins were used in the EMSAs as depicted.
DISCUSSION
Nitrate metabolism and its regulation has been best studied in E. coli, wherein Nar proteins are produced during anaerobic growth by the Fnr transcriptional regulator (45) and by the action of NarX/NarL and NarQ/NarP protein pairs that regulate transcription of nitrate reductase and nitrite export in response to nitrate/nitrite levels (28, 46). Based on the overall amino acid similarity between the E. coli and M. tuberculosis NarL proteins, a role for M. tuberculosis NarL (Rv0844c) in the regulation of nitrate/nitrite metabolism has long been proposed; however, no studies have elucidated the physiological function of the M. tuberculosis NarL protein. Given that mycobacteria can assimilate nitrate for growth (25, 47) and for anaerobic respiration (19) via reduction of nitrate to nitrite and, most importantly, that nitrate respiration plays a crucial role in mycobacterial persistence (6, 22, 27, 48), understanding the regulation of nitrate metabolism is of significant interest.
In this study, we focused on the role of the M. tuberculosis NarL response regulator and the underlying cognate and non-cognate signaling interactions that contribute to the regulation of nitrate metabolism during aerobic growth. We demonstrate that the Rv0845 gene encoding NarS, a putative histidine sensor kinase, interacts with Rv0844c, which encodes the NarL response regulator in vitro and in vivo and that NarS is the cognate sensor kinase for NarL. The NarL/NarS proteins constitute a functionally viable TCSS and display phosphotransfer kinetics typical of TCSSs. To gain insights into the regulatory role of NarL, a narL mutant strain of M. tuberculosis was constructed and characterized. Unlike E. coli, deletion of the narL gene in M. tuberculosis did not seem to affect nitrate reductase activity because nitrite generation in ΔnarL mutant was similar to H37Rv. However, transcription profiling of H37Rv and the ΔnarL mutant cultivated under conditions of aerobic growth (with or without nitrate) highlighted distinct changes in gene expression in the ΔnarL mutant grown with nitrate. We identified ∼30 genes that require NarL for expression in the presence of nitrate during aerobic growth. The narK2 gene encoding the NarK2 nitrate transporter protein (also regulated by the DevRS/DosT TCSS) was significantly down-regulated in the ΔnarL mutant. Interestingly, besides narK2, the overlap with the DevR regulon extended to 15 of 30 genes being common between the nitrate-responsive NarL and hypoxia-induced DevR-regulated gene sets. Notably, the overlap is also true for the DevR regulon induced by NO (8). This overlap together with expression analysis in M. tuberculosis H37Rv, ΔdevR, ΔdevSΔdosT, and ΔnarL mutant strains presented in Figs. 3–5 point toward co-regulatory roles of DevR and NarL RRs in the regulation of gene expression during aerobic nitrate metabolism. The analysis revealed that DevR is the primary RR regulating gene expression, but both NarL and DevR are required for optimum synergistic regulation. The inability of the NarL RR to directly bind to the promoter regions of target genes tested in this study is intriguing but does not preclude its binding to promoter regions of other members of the NarL regulon. The EMSA studies clearly show that NarL plays a supporting role; however, the extent of its contribution may vary among the genes common to NarL and DevR regulons, depending on their relative affinities for the target DNA. Moreover, co-regulation is evident only in the aerobic nitrate condition but is conspicuously missing in hypoxic conditions, suggesting that hypoxia is not likely to be the signal for NarL- and DevR-mediated co-regulation. It is conceivable that nitrate metabolic pathways, assimilatory or respiratory, are interconnected to allow rapid adaptation to subtle changes in the nitrogen and/or oxygen levels encountered by the bacteria during in vitro and in vivo growth. How the equilibrium of nitrate utilization and nitrite generation shifts during aerobic and hypoxic growth and its effects on temporal M. tuberculosis gene expression and physiology are not clear but are pertinent questions that will shed light on the regulatory function of NarL and its interplay with the DevRS/DosT TCSS.
It is important to state here that this is not the first report of regulation overlap with DevR regulon genes. So far, two M. tuberculosis RRs (PhoP and MprA) (49–52) and one Ser/Thr protein kinase (PknH) (53) have also been shown to activate the DevR regulon. However, to the best of our knowledge, this is the first report demonstrating activation of the DevRS/DosT TCSS during aerobic growth of M. tuberculosis in the presence of nitrate. From our results, it is evident that transcriptional activation of the Rv3134c-devR-devS operon is dependent on phosphorylation of DevR via the DevS/DosT sensor kinases even in aerobic nitrate/nitrite conditions. Using a nitrate metabolism and short-pulse model, we demonstrate that it is the endogenously produced nitrite that is the metabolic cue for induction of these genes. Because nitrite can be converted to NO (54, 55), our results are in accordance with the previous observations wherein low levels of NO activate the DevRS/DosT TCSS (8). The role of nitrite as a signal has been established for mammalian cells (56); however, in M. tuberculosis, its full potential in pathogenesis is relatively unexplored.
The interplay of the Nar and Dev/Dos systems reiterates the emerging hypothesis that mycobacteria execute coordinated signaling responses to facilitate metabolic adaptation during growth. Several questions arise regarding the underlying mechanisms of these co-regulatory responses and the nature of interactions. We addressed these questions on three separate levels by evaluating 1) cross-phosphorylation of DevR and NarL RRs by non-cognate sensor kinases, 2) DevR-NarL protein-protein interactions, and 3) DevR-NarL-DNA interactions. Our studies rule out any phosphorylation cross-talk between NarS HK and DevR RR. In agreement with a recent report (42), we observed that DevS can phosphorylate NarL but with very poor efficiency. The lack of in vivo interactions between NarL and DevS supports the notion that DevS-mediated NarL phosphorylation may not be relevant in vivo.
A novel finding of this study is that NarL and DevR RRs interact within mycobacteria and that these interactions can enable cooperative binding of both proteins to the target gene promoter regions for concerted regulation of gene expression. Although the exact mechanism of binding (e.g. whether NarL binding to DevR stabilizes the DevR-DNA interaction and the stoichiometry of these proteins) is not identified, it is clear that DevR activation is critical for cooperative action with NarL. Most RRs, including DevR, undergo homodimerization for downstream transcriptional regulation (57, 58). Although homodimers are more prevalent, formation of heterodimers cannot be completely excluded. In fact, recent evidence suggests the formation of heterodimers as an additional mechanism that facilitates cross-regulation and expansion of signaling networks (59, 60). We hypothesize that formation of NarL-DevR heterodimers may arise from high sequence similarities between them.
In summary, this study provides insights into the functionality of M. tuberculosis NarL/NarS TCSS and regulatory function of NarL protein and elucidates, for the first time, the underlying intricate signaling mechanisms that regulate nitrate metabolism in mycobacteria. The question regarding the biological relevance for the supporting role of NarL, however, still remains. The questions of whether NarL can co-regulate genes with other mycobacterial RR proteins and whether this can be another mechanism to integrate or fine tune signaling pathways in M. tuberculosis merit future investigations.
Supplementary Material
Acknowledgments
We acknowledge Genotypic Technology Pvt. Ltd. (Bangalore, India) for the microarray processing and data analysis and the Goldwater Environmental Laboratory (Arizona State University) for the flow injection analysis reported in this publication. We are grateful to Drs. David Sherman for the kind gift of M. tuberculosis ΔdosR and ΔdosSΔdosT mutant strains; Adrie Steyn for M-PFC vectors, pUAB200::devR, and pUAB200::devS; Jaya S. Tyagi for DevScyto, DosTcyto, and GST-DevR expression plasmids; and Michael Lin for pcDNA3-mRuby2. We thank James Megehee Jr. and Jiseon Yang for help with the ΔnarL mutant construction and M-PFC assay, respectively, and Mayooreshwar P. Rajankar (Indian Institute of Science) for generating Nar expression constructs.
This work was supported, in whole or in part, by National Institutes of Health Grant AI46428. This research was also supported by Department of Biotechnology, India, Grant BT/PR15001/BRB/10/897; Council of Scientific and Industrial Research, India, Grant-in-aid 37 (1435) 10-EMR-II (to D. K. S.); and Arizona State University start-up funding (to J. E. C.-C.).
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE59063).

This article contains supplemental data sets S1–S3.
- TCSS
- two-component signal transduction system
- HK
- histidine kinase
- M-PFC
- mycobacterial protein fragment complementation assay
- mDHFR
- murine dihydrofolate reductase
- RR
- response regulator
- SPR
- surface plasmon resonance
- TRIM
- trimethoprim
- qRT-PCR
- quantitative RT-PCR.
REFERENCES
- 1. Chakraborti P. K., Matange N., Nandicoori V. K., Singh Y., Tyagi J. S., Visweswariah S. S. (2011) Signalling mechanisms in Mycobacteria. Tuberculosis 91, 432–440 [DOI] [PubMed] [Google Scholar]
- 2. Haydel S. E., Clark-Curtiss J. E. (2004) Global expression analysis of two-component system regulator genes during Mycobacterium tuberculosis growth in human macrophages. FEMS Microbiol. Lett. 236, 341–347 [DOI] [PubMed] [Google Scholar]
- 3. Zahrt T. C., Deretic V. (2000) An essential two-component signal transduction system in Mycobacterium tuberculosis. J. Bacteriol. 182, 3832–3838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Haydel S. E., Malhotra V., Cornelison G. L., Clark-Curtiss J. E. (2012) The prrAB two-component system is essential for Mycobacterium tuberculosis viability and is induced under nitrogen-limiting conditions. J. Bacteriol. 194, 354–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bretl D. J., Demetriadou C., Zahrt T. C. (2011) Adaptation to environmental stimuli within the host: two-component signal transduction systems of Mycobacterium tuberculosis. Microbiol. Mol. Biol. Rev. 75, 566–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wayne L. G., Sohaskey C. D. (2001) Nonreplicating persistence of Mycobacterium tuberculosis. Annu Rev. Microbiol. 55, 139–163 [DOI] [PubMed] [Google Scholar]
- 7. Schnappinger D., Ehrt S., Voskuil M. I., Liu Y., Mangan J. A., Monahan I. M., Dolganov G., Efron B., Butcher P. D., Nathan C., Schoolnik G. K. (2003) Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198, 693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Voskuil M. I., Schnappinger D., Visconti K. C., Harrell M. I., Dolganov G. M., Sherman D. R., Schoolnik G. K. (2003) Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198, 705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Kumar A., Deshane J. S., Crossman D. K., Bolisetty S., Yan B. S., Kramnik I., Agarwal A., Steyn A. J. (2008) Heme oxygenase-1-derived carbon monoxide induces the Mycobacterium tuberculosis dormancy regulon. J. Biol. Chem. 283, 18032–18039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shiloh M. U., Manzanillo P., Cox J. S. (2008) Mycobacterium tuberculosis senses host-derived carbon monoxide during macrophage infection. Cell Host Microbe 3, 323–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Taneja N. K., Dhingra S., Mittal A., Naresh M., Tyagi J. S. (2010) Mycobacterium tuberculosis transcriptional adaptation, growth arrest and dormancy phenotype development is triggered by vitamin C. PLoS One 5, e10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sherman D. R., Voskuil M., Schnappinger D., Liao R., Harrell M. I., Schoolnik G. K. (2001) Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding α-crystallin. Proc. Natl. Acad. Sci. U.S.A. 98, 7534–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Park H. D., Guinn K. M., Harrell M. I., Liao R., Voskuil M. I., Tompa M., Schoolnik G. K., Sherman D. R. (2003) Rv3133c/dosR is a transcription factor that mediates the hypoxic response of Mycobacterium tuberculosis. Mol. Microbiol. 48, 833–843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Leistikow R. L., Morton R. A., Bartek I. L., Frimpong I., Wagner K., Voskuil M. I. (2010) The Mycobacterium tuberculosis DosR regulon assists in metabolic homeostasis and enables rapid recovery from nonrespiring dormancy. J. Bacteriol. 192, 1662–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Aly S., Wagner K., Keller C., Malm S., Malzan A., Brandau S., Bange F. C., Ehlers S. (2006) Oxygen status of lung granulomas in Mycobacterium tuberculosis-infected mice. J. Pathol. 210, 298–305 [DOI] [PubMed] [Google Scholar]
- 16. Barry C. E., 3rd, Boshoff H. I., Dartois V., Dick T., Ehrt S., Flynn J., Schnappinger D., Wilkinson R. J., Young D. (2009) The spectrum of latent tuberculosis: rethinking the biology and intervention strategies. Nat. Rev. Microbiol. 7, 845–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Loebel R. O., Shorr E., Richardson H. B. (1933) The influence of adverse conditions upon the respiratory metabolism and growth of human tubercle bacilli. J. Bacteriol. 26, 167–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Via L. E., Lin P. L., Ray S. M., Carrillo J., Allen S. S., Eum S. Y., Taylor K., Klein E., Manjunatha U., Gonzales J., Lee E. G., Park S. K., Raleigh J. A., Cho S. N., McMurray D. N., Flynn J. L., Barry C. E., 3rd. (2008) Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect. Immun. 76, 2333–2340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Shi L., Sohaskey C. D., Kana B. D., Dawes S., North R. J., Mizrahi V., Gennaro M. L. (2005) Changes in energy metabolism of Mycobacterium tuberculosis in mouse lung and under in vitro conditions affecting aerobic respiration. Proc. Natl. Acad. Sci. U.S.A. 102, 15629–15634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hedgecock L. W., Costello R. L. (1962) Utilization of nitrate by pathogenic and saprophytic mycobacteria. J. Bacteriol. 84, 195–205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Tan M. P., Sequeira P., Lin W. W., Phong W. Y., Cliff P., Ng S. H., Lee B. H., Camacho L., Schnappinger D., Ehrt S., Dick T., Pethe K., Alonso S. (2010) Nitrate respiration protects hypoxic Mycobacterium tuberculosis against acid- and reactive nitrogen species stresses. PLoS One 5, e13356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sohaskey C. D. (2008) Nitrate enhances the survival of Mycobacterium tuberculosis during inhibition of respiration. J. Bacteriol. 190, 2981–2986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cunningham-Bussel A., Zhang T., Nathan C. F. (2013) Nitrite produced by Mycobacterium tuberculosis in human macrophages in physiologic oxygen impacts bacterial ATP consumption and gene expression. Proc. Natl. Acad. Sci. U.S.A. 110, E4256–E4265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bogdan C. (2001) Nitric oxide and the immune response. Nat. Immunol. 2, 907–916 [DOI] [PubMed] [Google Scholar]
- 25. Malm S., Tiffert Y., Micklinghoff J., Schultze S., Joost I., Weber I., Horst S., Ackermann B., Schmidt M., Wohlleben W., Ehlers S., Geffers R., Reuther J., Bange F. C. (2009) The roles of the nitrate reductase NarGHJI, the nitrite reductase NirBD and the response regulator GlnR in nitrate assimilation of Mycobacterium tuberculosis. Microbiology 155, 1332–1339 [DOI] [PubMed] [Google Scholar]
- 26. Cole S. T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S. V., Eiglmeier K., Gas S., Barry C. E., 3rd, Tekaia F., Badcock K., Basham D., Brown D., Chillingworth T., Connor R., Davies R., Devlin K., Feltwell T., Gentles S., Hamlin N., Holroyd S., Hornsby T., Jagels K., Krogh A., McLean J., Moule S., Murphy L., Oliver K., Osborne J., Quail M. A., Rajandream M. A., Rogers J., Rutter S., Seeger K., Skelton J., Squares R., Squares S., Sulston J. E., Taylor K., Whitehead S., Barrell B. G. (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393, 537–544 [DOI] [PubMed] [Google Scholar]
- 27. Khan A., Sarkar D. (2012) Nitrate reduction pathways in mycobacteria and their implications during latency. Microbiology 158, 301–307 [DOI] [PubMed] [Google Scholar]
- 28. Rabin R. S., Stewart V. (1993) Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate- and nitrite-regulated gene expression in Escherichia coli K-12. J. Bacteriol. 175, 3259–3268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Saini D. K., Malhotra V., Dey D., Pant N., Das T. K., Tyagi J. S. (2004) DevR-DevS is a bona fide two-component system of Mycobacterium tuberculosis that is hypoxia-responsive in the absence of the DNA-binding domain of DevR. Microbiology 150, 865–875 [DOI] [PubMed] [Google Scholar]
- 30. Lam A. J., St-Pierre F., Gong Y., Marshall J. D., Cranfill P. J., Baird M. A., McKeown M. R., Wiedenmann J., Davidson M. W., Schnitzer M. J., Tsien R. Y., Lin M. Z. (2012) Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods 9, 1005–1012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Singh A., Mai D., Kumar A., Steyn A. J. (2006) Dissecting virulence pathways of Mycobacterium tuberculosis through protein-protein association. Proc. Natl. Acad. Sci. U.S.A. 103, 11346–11351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bardarov S., Bardarov S., Jr., Pavelka M. S., Jr., Sambandamurthy V., Larsen M., Tufariello J., Chan J., Hatfull G., Jacobs W. R., Jr. (2002) Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148, 3007–3017 [DOI] [PubMed] [Google Scholar]
- 33. Malhotra V., Tyagi J. S., Clark-Curtiss J. E. (2009) DevR-mediated adaptive response in Mycobacterium tuberculosis H37Ra: links to asparagine metabolism. Tuberculosis 89, 169–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Malhotra V., Okon B. P., Clark-Curtiss J. E. (2012) Mycobacterium tuberculosis protein kinase K enables growth adaptation through translation control. J. Bacteriol. 194, 4184–4196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Parish T., Smith D. A., Kendall S., Casali N., Bancroft G. J., Stoker N. G. (2003) Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71, 1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Laub M. T., Biondi E. G., Skerker J. M. (2007) Phosphotransfer profiling: systematic mapping of two-component signal transduction pathways and phosphorelays. Methods Enzymol. 423, 531–548 [DOI] [PubMed] [Google Scholar]
- 37. Agrawal R., Saini D. K. (2014) Rv1027c-Rv1028c encode functional KdpDE two-component system in Mycobacterium tuberculosis. Biochem. Biophys. Res. Commun. 446, 1172–1178 [DOI] [PubMed] [Google Scholar]
- 38. Bhattacharya M., Biswas A., Das A. K. (2010) Interaction analysis of TcrX/Y two component system from Mycobacterium tuberculosis. Biochimie 92, 263–272 [DOI] [PubMed] [Google Scholar]
- 39. Stock A. M., Robinson V. L., Goudreau P. N. (2000) Two-component signal transduction. Annu. Rev. Biochem. 69, 183–215 [DOI] [PubMed] [Google Scholar]
- 40. Roberts D. M., Liao R. P., Wisedchaisri G., Hol W. G., Sherman D. R. (2004) Two sensor kinases contribute to the hypoxic response of Mycobacterium tuberculosis. J. Biol. Chem. 279, 23082–23087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Chauhan S., Tyagi J. S. (2008) Cooperative binding of phosphorylated DevR to upstream sites is necessary and sufficient for activation of the Rv3134c-devRS operon in Mycobacterium tuberculosis: implication in the induction of DevR target genes. J. Bacteriol. 190, 4301–4312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cho H. Y., Kang B. S. (2014) Serine 83 in DosR, a response regulator from Mycobacterium tuberculosis, promotes its transition from an activated, phosphorylated state to an inactive, unphosphorylated state. Biochem. Biophys. Res. Commun. 444, 651–655 [DOI] [PubMed] [Google Scholar]
- 43. Chauhan S., Sharma D., Singh A., Surolia A., Tyagi J. S. (2011) Comprehensive insights into Mycobacterium tuberculosis DevR (DosR) regulon activation switch. Nucleic Acid Res. 39, 7400–7414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Gautam U. S., Chauhan S., Tyagi J. S. (2011) Determinants outside the DevR C-terminal domain are essential for cooperativity and robust activation of dormancy genes in Mycobacterium tuberculosis. PLoS One 6, e16500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Darwin A. J., Ziegelhoffer E. C., Kiley P. J., Stewart V. (1998) Fnr, NarP, and NarL regulation of Escherichia coli K-12 napF (periplasmic nitrate reductase) operon transcription in vitro. J. Bacteriol. 180, 4192–4198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Li J., Kustu S., Stewart V. (1994) In vitro interaction of nitrate-responsive regulatory protein NarL with DNA target sequences in the fdnG, narG, narK and frdA operon control regions of Escherichia coli K-12. J. Mol. Biol. 241, 150–165 [DOI] [PubMed] [Google Scholar]
- 47. Khan A., Akhtar S., Ahmad J. N., Sarkar D. (2008) Presence of a functional nitrate assimilation pathway in Mycobacterium smegmatis. Microb. Pathog. 44, 71–77 [DOI] [PubMed] [Google Scholar]
- 48. Höner zu Bentrup K., Russell D. G. (2001) Mycobacterial persistence: adaptation to a changing environment. Trends Microbiol. 9, 597–605 [DOI] [PubMed] [Google Scholar]
- 49. Bretl D. J., He H., Demetriadou C., White M. J., Penoske R. M., Salzman N. H., Zahrt T. C. (2012) MprA and DosR coregulate a Mycobacterium tuberculosis virulence operon encoding Rv1813c and Rv1812c. Infect. Immun. 80, 3018–3033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Gonzalo-Asensio J., Mostowy S., Harders-Westerveen J., Huygen K., Hernández-Pando R., Thole J., Behr M., Gicquel B., Martín C. (2008) PhoP: a missing piece in the intricate puzzle of Mycobacterium tuberculosis virulence. PLoS One 3, e3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. He H., Bretl D. J., Penoske R. M., Anderson D. M., Zahrt T. C. (2011) Components of the Rv0081-Rv0088 locus, which encodes a predicted formate hydrogenlyase complex, are coregulated by Rv0081, MprA, and DosR in Mycobacterium tuberculosis. J. Bacteriol. 193, 5105–5118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pang X., Vu P., Byrd T. F., Ghanny S., Soteropoulos P., Mukamolova G. V., Wu S., Samten B., Howard S. T. (2007) Evidence for complex interactions of stress-associated regulons in an mprAB deletion mutant of Mycobacterium tuberculosis. Microbiology 153, 1229–1242 [DOI] [PubMed] [Google Scholar]
- 53. Chao J. D., Papavinasasundaram K. G., Zheng X., Chávez-Steenbock A., Wang X., Lee G. Q., Av-Gay Y. (2010) Convergence of Ser/Thr and two-component signaling to coordinate expression of the dormancy regulon in Mycobacterium tuberculosis. J. Biol. Chem. 285, 29239–29246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhang Z., Naughton D., Winyard P. G., Benjamin N., Blake D. R., Symons M. C. (1998) Generation of nitric oxide by a nitrite reductase activity of xanthine oxidase: a potential pathway for nitric oxide formation in the absence of nitric oxide synthase activity. Biochem. Biophys. Res. Commun. 249, 767–772 [DOI] [PubMed] [Google Scholar]
- 55. Shiva S. (2013) Nitrite: a physiological store of nitric oxide and modulator of mitochondrial function. Redox Biol. 1, 40–44 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bryan N. S., Fernandez B. O., Bauer S. M., Garcia-Saura M. F., Milsom A. B., Rassaf T., Maloney R. E., Bharti A., Rodriguez J., Feelisch M. (2005) Nitrite is a signaling molecule and regulator of gene expression in mammalian tissues. Nat. Chem. Biol. 1, 290–297 [DOI] [PubMed] [Google Scholar]
- 57. Wisedchaisri G., Wu M., Rice A. E., Roberts D. M., Sherman D. R., Hol W. G. (2005) Structures of Mycobacterium tuberculosis DosR and DosR-DNA complex involved in gene activation during adaptation to hypoxic latency. J. Mol. Biol. 354, 630–641 [DOI] [PubMed] [Google Scholar]
- 58. Wisedchaisri G., Wu M., Sherman D. R., Hol W. G. (2008) Crystal structures of the response regulator DosR from Mycobacterium tuberculosis suggest a helix rearrangement mechanism for phosphorylation activation. J. Mol. Biol. 378, 227–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Gao R., Tao Y., Stock A. M. (2008) System-level mapping of Escherichia coli response regulator dimerization with FRET hybrids. Mol. Microbiol. 69, 1358–1372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Al-Bassam M. M., Bibb M. J., Bush M. J., Chandra G., Buttner M. J. (2014) Response regulator heterodimer formation controls a key stage in Streptomyces development. PLoS Genet. 10, e1004554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Saini D. K., Pant N., Das T. K., Tyagi J. S. (2002) Cloning, overexpression, purification, and matrix-assisted refolding of DevS (Rv 3132c) histidine protein kinase of Mycobacterium tuberculosis. Protein Expr. Purif. 25, 203–208 [DOI] [PubMed] [Google Scholar]
- 62. Saini D. K., Malhotra V., Tyagi J. S. (2004) Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett. 565, 75–80 [DOI] [PubMed] [Google Scholar]
- 63. Bagchi G., Chauhan S., Sharma D., Tyagi J. S. (2005) Transcription and autoregulation of the Rv3134c-devR-devS operon of Mycobacterium tuberculosis. Microbiology 151, 4045–4053 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.