Figure 2.
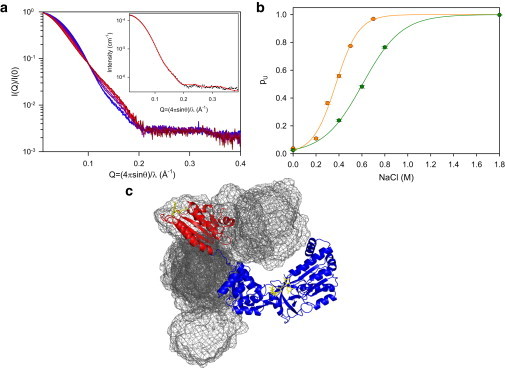
SAXS analysis of the ionic strength-mediated conformational transition of CPR. (a) Scattering curves from 0 M (blue) to 0.7 M (red) NaCl concentration at pH 7.4, normalized to protein concentration. (Dark red) Extrapolated curve at high ionic strength. (Inset) Fit of the 0 M scattering curve to the crystal structure of the closed conformation (6) with loop conformation optimization using DADIMODO software (28). (b) Fractions of the U state (pU) at pH 7.4 (orange) and pH 6.7 (green), with their respective fits to the denaturation law. (c) One of the 11 EOM-derived structures used to reproduce the scattering curve extrapolated for the U state is represented as ribbons (blue, FAD/connecting domain; red, FMN domain). The mesh (calculated with the MESH feature of the PYMOL software (42)) represents the van der Waals surface of the reunion of the 11 FMN domains determined by EOM. All conformations were superimposed on the FAD/connecting domain. To see this figure in color, go online.
