Figure 2.
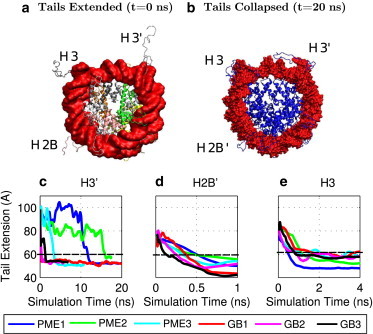
Three sets of independent MD simulations of the nucleosome tail collapse. (a) PDB structure with three histone tails (H3, H3′, and H2B′) extended. (b) Structure with the positively charged histone tails collapsed onto the negatively charged DNA. DNA surface is shown in red and the histone backbone structure is shown in blue. Tail extension is measured as the distance from the N-terminus tail to the center of geometry of the DNA. (c–e) Results show moving average values, averaged over 0.5 ns, from three explicit-solvent (TIP3P) PME and GB simulations. Connecting lines are shown to guide the eye. Images were rendered using VMD (100). To see this figure in color, go online.
