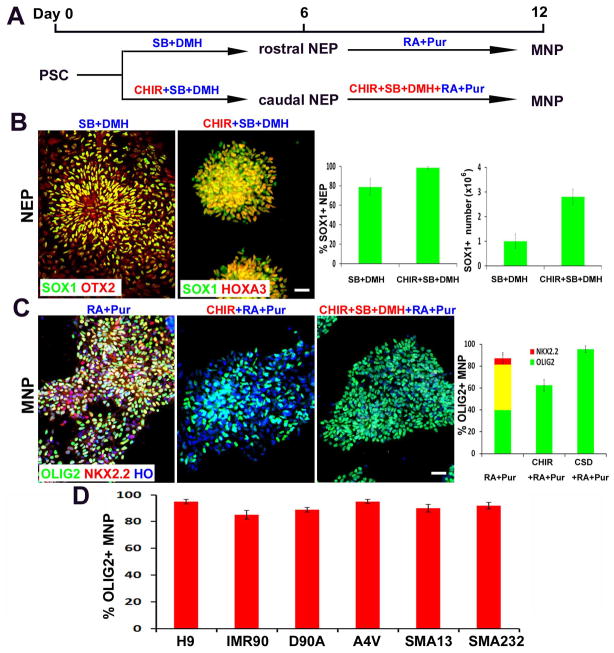
Figure 1. Generation of highly-pure population of MNPs from hPSCs.
(A) Schematics showing the time course and small molecule cocktail for hPSC differentiation into MNPs. (B) Representative images of SOX1+ NEPs after 6 days of culture in CHIR+SB+DMH vs. SB+DMH condition. The regional identity (OTX2+ vs. HOXA3+) were stained. Scale bars: 50μm. Quantification of SOX1+ NEP percentage and number is shown on right (>500 cells from random fields were manually counted in each condition). The bar graph shows the mean±s.d. (n=3 in each condition). (C) Representative images of pure MNPs at Day12 under different conditions, which express OLIG2 (green) but not NKX2.2 (red). Scale bars: 50μm. Quantification of OLIG2+, NKX2.2+ and OLIG2+/NKX2.2+ cells is shown on right (>500 cells from random fields were manually counted in each condition). The bar graph shows the mean±s.d. (n=3 in each condition). (D) The efficiency of OLIG2+ MNP differentiation from multiple hPSC lines (>500 cells from random fields were manually counted in each cell line). The bar graph shows the mean±s.d. (n=3 in each cell line).