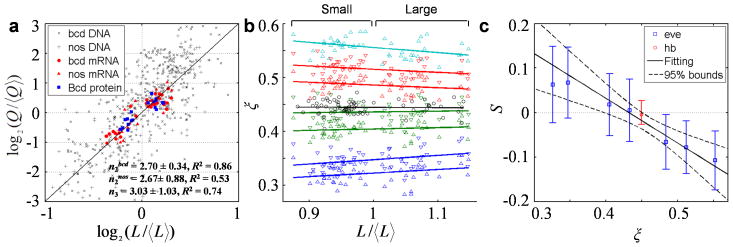
Fig. 5. Scaling power measurements in embryos and evaluations of model predictions.
a) Superimposed plots showing scaling power measurements in the two inbred lines; also shown is a reference line with a slope of 3. Datapoints for bcd DNA, nos DNA, bcd mRNA, nos mRNA, and Bcd protein are shown as x marks, crosses, circles, diamonds, and squares, respectively. b) Scatter plot of relative positions of hb (black) or the anterior 7 eve (colored) expression boundaries in embryos from these inbred lines (see Supplementary Fig. 7 for a display showing all eve boundaries). Color code for eve data: blue, green, red, and magenta for expression stripes 1–4, respectively, with upright and downward triangles representing anterior and posterior boundaries, respectively. c) Measured values of scaling coefficient S, estimated as the slope of a regression line (solid lines in b), are plotted as a function of boundary position ξ. Error bars represent 95% CI of each fitted slope. Results for hb and eve are shown in red and blue, respectively. Solid line shown is a linear fit for the selected boundaries (see text; R2 = 0.94), and dashed lines are 95% prediction bounds of this fit. In another linear fit using boundaries within a wider range of the embryo (hb and the 1st through 7th of eve), we obtained consistent estimates (ξC = 0.422 and nA = 2.13) that likely have also incorporated the impact of terminal system inputs on boundaries that are closer to the anterior pole 24,51. Extended discussion about scaling coefficient S: Under its current definition (see text), S = 0 denotes perfect scaling of a gene’s expression boundary. If S < 0 or S > 0, the boundary is either under- or over-scaled, respectively. This evaluation is consistent with a previous analysis 16 performed under the framework of a differently defined scaling coefficient SBerg, as dictated by the relationship S = ξ (SBerg −1).