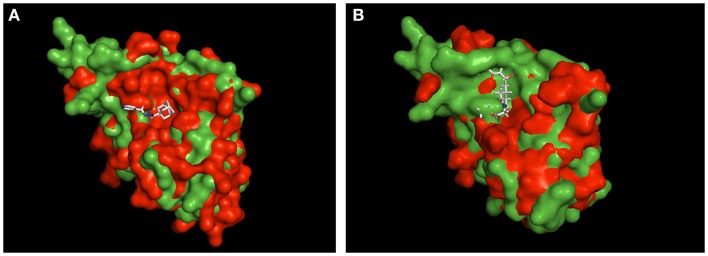
Figure 3.
Structural alignment of Mip to known PPIase-inhibitor complexes. Shown are the structural overlays of the surface model of Mip (1FD9, green) with (A) the PvFKBP35-SAR-complex (4MVG, red) and (B) the BpML1- cycloheximide N-ethylethanoate complex (2KO7, red). The ligands are shown as sticks. The overlay in (A) suggests that Mip and PvFKBP35 are topologically more similar in the hydrophobic cleft, where SAR docks. Accordingly, a comparable binding mode of MT_30.32 or MT_30.51 to Mip via their adamantyl moiety can be assumed. The overlay in (B) reveals substantial topological differences between Mip and BpML1 in the loop region and the vicinity of the hydrophobic cleft that are reported to interact with cycloheximide N-ethylethanoate. Hence, the cycloheximide portions of MT_30.32 and MT_30.51 most probably bind in a different mode to Mip. The structural alignments were performed using PyMOL Molecular Graphics System, Version 1.5.0.4 Schrödinger, LLC.