Figure 4.
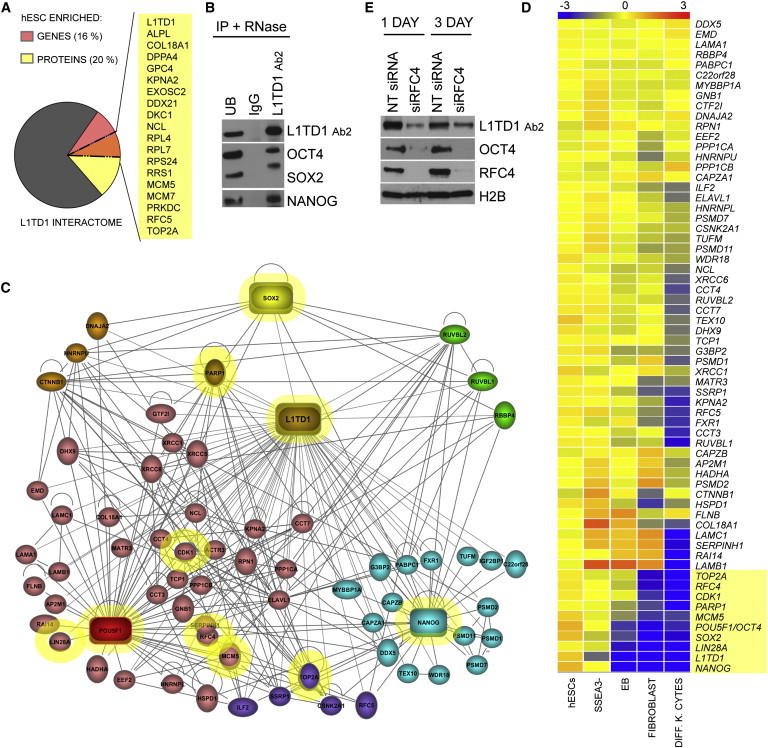
L1TD1 Connects Uncharacterized Factors to Core Pluripotency Regulation
(A) hESC-enriched genes (Assou et al., 2007) and proteins (Jadaliha et al., 2012; Van Hoof et al., 2006) in the L1TD1 interactome. Overlapping hits are listed.
(B) WB detection of OCT4, SOX2, and NANOG in L1TD1, IgG control, and UB IP reactions. hESC line HS360.
(C) Proteins that interact with L1TD1 and OCT4 (n = 45), NANOG (n = 25), or SOX2 (n = 8) (Ding et al., 2012; Nitzsche et al., 2011; van den Berg et al., 2010). The figure was constructed with the use of IPA software.
(D) RNA expression of proteins represented in Figure 4C in hESCs and differentiated cells. EB, embryoid body; DIFF.K.CYTES, differentiated keratinocytes. Microarray data collected from previous studies (Enver et al., 2005; Golan-Mashiach et al., 2005; Skottman et al., 2005). The figure was generated from normalized raw files using GeneSpring GX Software (Agilent Technologies Genomics). The top ten differentiation-responsive genes are highlighted in yellow.
(E) Effect of RFC4 silencing (siRNA1) on the expression of L1TD1 and OCT4. hESC line HS360. Quantifications can be found in the Supplemental Results.
See also Figure S2.
