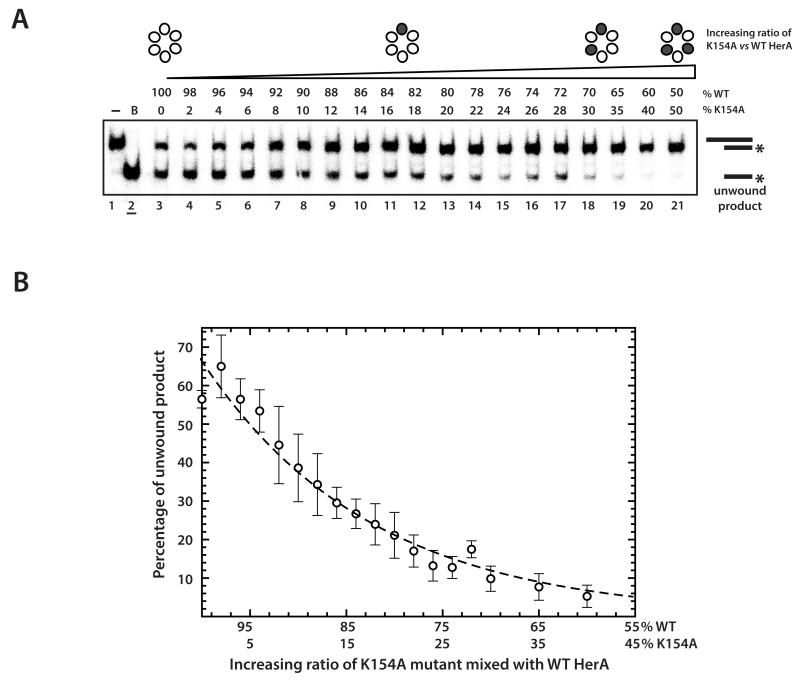
Figure 7. Mutant doping assays to examine the ordering of catalytic events around the hexameric ring.
(A) Mutant doping with the K154A HerA in a DNA unwinding assay. Lane 1, no protein control; lane 2 (underlined), boiled (unwound) control; lanes 3-21, native (WT) HerA and an increasing proportion of K154A HerA (0 to 50% K154A in 2% increments, respectively). The HerA proteins were mixed at 60°C, prior to the addition of D58A (nuclease inactivated) NurA at 60°C to form the HerA-NurA complex. After addition of the DNA substrate (32P radiolabelled on one strand, as denoted by the asterisk) the reactions were terminated after 30 minutes at 60°C by the addition of SDS and EDTA. Products were resolved by 12% PAGE and visualised by phosphoimaging. (B) Quantification and fitting of the mutant doping data in the unwinding assays shown in (A). The data points represent the mean of three independent repeats and the error bars one standard deviation. The data was fitted to a first order exponential decay model using ‘ProFit’ software (www.quansoft.com).