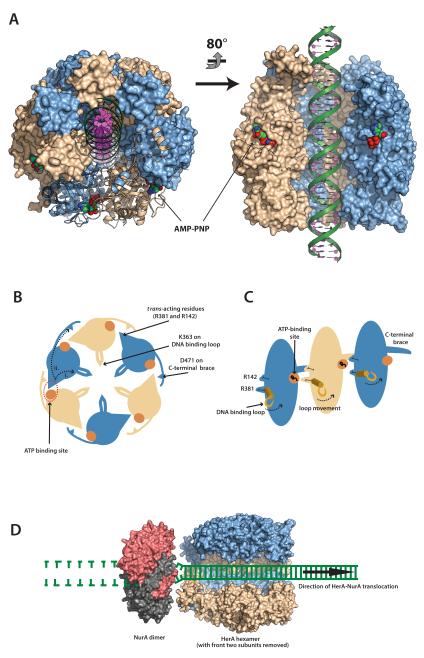
Figure 9. A model for dsDNA processing by the HerA-NurA complex.
(A) 40 bp dsDNA modelled into the DNA-binding channel of the HerA hexamer, coloured as in Fig. 1C. AMP-PNP molecules are shown as spacefill models. Left; Top-down view of the HerA hexamer tilted slightly from the vertical position, with four adjacent protomers shown in surface representation and two shown as ribbons. Right; Side-view of the HerA hexamer (HAS-barrels at top) with two front protomers omitted. Modelled dsDNA is threaded through the DNA-binding channel of HerA. Models generated using PyMOL (57). (B) Schematic representation of the HerA hexameric ring viewed in the plane of DNA binding. Alternating subunits are coloured in light blue and wheat, respectively. The nucleotide binding sites, indicated by orange circles, interact in trans with the adjacent subunit via the arginine residues R142 and R381. The putative DNA-binding loops are indicated within the central cavity. The C-terminal “brace” is also presented. The conserved D471 residue on this extension interacts with the trans-acting R142 residue of the neighbouring subunit. Movements of the R381 trans-acting arginine finger will likely translate to the DNA-binding loops and coordinate translocation (i), while communication between the subunits might be regulated by the D471 brace aspartate (ii). (C) Cutaway representation of three HerA subunits, viewed perpendicular to the plane of DNA binding. The association of the D471 C-terminal brace aspartate with the R142 trans-acting arginine is also illustrated. (D) A model for how HerA and NurA might cooperate to process DNA ends. HerA (PDB id 4D2I) is viewed from the side, with two subunits removed to reveal the DNA-binding channel. Interaction with dimeric NurA (red and grey, PDB id 2YGK) is mediated via the alpha-helical extensions at the N-terminal HAS-barrels of HerA, and reciprocal hydrophobic surfaces on the NurA annulus (16). Double-stranded DNA is channelled through HerA and unwound by the ploughshare action of NurA. In the model, both DNA strands are engaged by the NurA dimer, resulting in wholesale degradation of the substrate.