Figure 3.
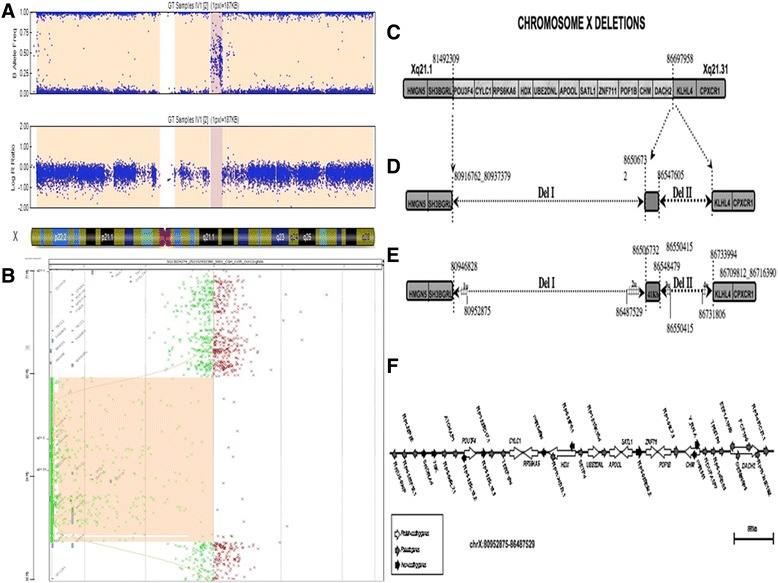
Info on deleted region. A) Graph of the marker-specific fluorescence (logR) and allele-specific fluorescence of SNPs on chromosome X in 1 of the patients. The deleted region is marked in dark pink. B) A-CGH profile of chromosome from the Agilent 1x1M array in the subject IV-2 shown that the deletion of 5,77 Mbp includes a small undeleted region of about 40 Kb. C) SNPs array results: deleted genes are reported in the light grey zone and undeleted genes in the dark grey zone. D) CGH array result: zones deleted are represented as dotted arrows. E) Refinement of breakpoint deletions results by PCR analysis. Gridded zone represents regions that remained undetermined. F) Schematic representation of different classes of genes identified in silico in the deleted region.The scale bar in the panel F refers to the entire genomic region that is deleted, whereas single genes or transcripts are not in scale. However, the genomic localization of each gene in this region is respected.
