Figure 5.
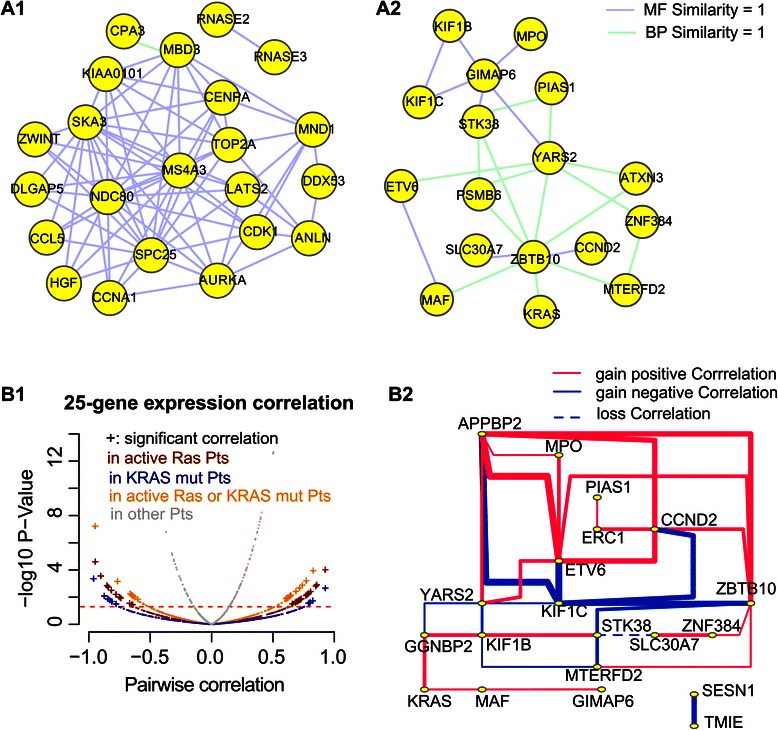
The computationally evaluated biological relevance of genes in the identified gene-sets. Panel A) Genes in the identified cluster of gene-sets share common functions. Sub-panel 1 represents the 30 LSC- representative genes and sub-panel 2 represents 25 LSC+ representative genes. Gene Ontology semantic similarity analysis reveals that 2/3 of the genes share the same function (the Lin distance =1). Line color codes the molecular function and biological process respectively. Panel B) Volcano plot (subpanel 1) of pairwise correlation tests between any two LSC+ 25-gene members (TCAG data). There are 31 significant co-expressions across 10 AML patients who show positive RAS activity (dark red). This co-expression disappears among the other 187 patients (grey), the 7 patients who carry somatic mutations of KRAS genes (blue), and the 16 patients carrying positive RAS activity or somatic RAS mutation (orange). Subpanel 2 illustrates the RAS activity-dependent co-expressed gene-pairs in a network. Comparing patients showing active molecular activity of RAS with patients showing normal RAS status, 31 pairs of genes gain significant co-expression, including 20 out of the 25 LSC+ representative genes (solid lines), whereas 1 gene-pair loses its co-expression in normal RAS status (the dashed line). Red lines represent positive correlations and blue lines represent negative correlations. Line widths correspond to Spearman correlation coefficients.
