Figure 1. The ‘nascent transcript’ model and a self-reinforcing epigenetic loop in S. pombe.
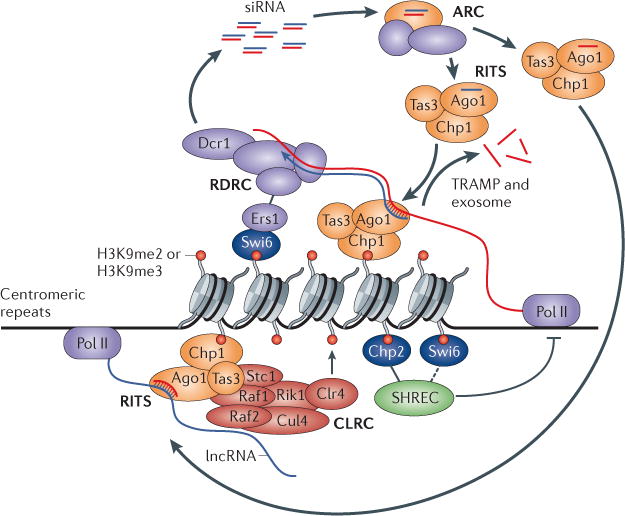
In Schizosaccharomyces pombe, the RNA-induced transcriptional silencing (RITS) complex establishes a physical connection between small interfering RNAs (siRNAs) and heterochromatin by targeting a nascent transcript, and forms the basis of a self-sustaining feedback mechanism that couples siRNA production to chromatin modification. A siRNA-targeted centromeric long non-coding (lncRNA) bound to the RITS complex becomes a template for double-stranded RNA (dsRNA) synthesis by the RNA-dependent RNA polymerase complex (RDRC, which is composed of Rdp1, Hrr1 and Cid12) and generation by Dicer 1 (Dcr1) of new siRNAs, leading to further targeting of the RITS complex after passage of Argonaute (Ago) through the ARC (Ago siRNA chaperone) complex. The Chp1 subunit of the RITS complex anchors the complex onto nucleosomes with histone H3 lysine 9 (H3K9) methylation, and the RITS complex recruits the Clr4–Rik1–Cul4 (CLRC, of which Clr4 is the methyltransferase) complex via Rik1 and Stc1 to promote the further spread of H3K9 methylation. The heterochromatin protein 1 (HP1) homologue Swi6 binds to methylated H3K9 and promotes RDRC recruitment and siRNA biogenesis via the silencing factor Ers1. Swi6, and particularly the other HP1 protein Chp2, help to restrict RNA polymerase II (Pol II) access by recruiting the Snf2–histone deacetylase repressor complex (SHREC). The TRAMP non-canonical poly(A) polymerase and the exosome also contribute to silencing. Together, the RITS complex and the nascent lncRNA transcript provide a hub for the assembly of machineries that make siRNAs, modify histones and silence gene expression. me2, dimethylation.
