Abstract
Neuroblastoma (NB) is a childhood malignant tumor that arises from precursor cells of the sympathetic nervous system. Spontaneous regression is a phenomenon unique to NBs and is caused by differentiation of tumor cells. PES1 is a multifunctional protein with roles in both neural development and ribosome biogenesis. Various kinds of models have revealed the significance of PES1 in neurodevelopment. However, the roles of PES1 in NB tumorigenesis and differentiation have remained unknown. Here we show that NB cases with MYCN amplification and clinically unfavorable stage (INSS stage 4) express higher levels of PES1. High PES1 expression was associated with worse overall and relapse-free survival. In NB cell lines, PES1 knockdown suppressed tumor cell growth and induced apoptosis. This growth inhibition was associated with the expression of NB differentiation markers. However, when the differentiation of NB cell lines was induced by the use of all-trans retinoic acid, there was a corresponding decrease in PES1 expression. Pes1 expression of tumorspheres originated from MYCN transgenic mice also diminished after the induction of differentiation with growth factors. We also reanalyzed the distribution of PES1 in the nucleolus. PES1 was localized in the dense fibrillar component, but not in the granular component of nucleoli. After treatment with the DNA-damaging agent camptothecin, this distribution was dramatically changed to diffuse nucleoplasmic. These data suggest that PES1 is a marker of NB outcome, that it regulates NB cell proliferation, and is associated with NB differentiation.
Keywords: Differentiation, neuroblastoma, nucleolus, PES1, tumorigenesis
Neuroblastoma (NB) is the most common extracranial solid malignant tumor of childhood and is derived from precursor cells of the sympathetic nervous system.1 Unlike in the case of other malignant tumors, a subset of patients with NBs show spontaneous regression without any treatment. This phenomenon is caused by differentiation and programmed cell death of tumor cells.2 Differentiation induction and proliferation inhibition with 13-cis-retinoic acid is thus one of the effective therapies for high-risk NBs.3 MYCN is a well-known driver gene of NBs. Twenty to thirty percent of NB cases show MYCN amplification and poor prognosis.4 Although MYCN, which has a helix–loop–helix domain and belongs to the MYC family of transcription factors, activates the transcription of various kinds of growth signals, recent data show that MYCN is also required for the differentiation program.5
The gene PES1 (and its zebrafish ortholog pes) is highly conserved from yeast to humans. PES1 was originally identified in zebrafish in an insertional mutagenesis screen. PES1 mutant embryos exhibited brains with reduced volumes and other developmental abnormalities.6 PES1 mutant overexpression disrupted oligodendrocyte development, oligodendrocyte migration, and axon extension.7 In the developing Xenopus laevis, pes1 is expressed in the migrating cranial neural crest cells. Inhibition of pes1 using morpholino oligonucleotide caused neural crest migration impairment.8 In developing mice, Pes1 displayed a distinct spatial and temporal pattern of gene expression with high levels in the germinal zone and other specific brain regions that contain neural progenitor cells and postmitotic neurons.9 In addition to these neurodevelopmental roles, PES1 Plays a crucial part in ribosome neogenesis,10,11 particularly in pre-ribosomal RNA (pre-rRNA) processing. In interphase cells, PES1 is localized in the nucleolus, an intranuclear body where ribosome neogenesis takes place.12,13 Previous reports revealed that PES1 was upregulated in glioblastoma, breast cancer, and head and neck squamous cell carcinoma.9,14,15 The close relationship between ribosome biogenesis and carcinogenesis could be the reason for these upregulations. PES1 knockdown has been shown to inhibit the proliferation and tumorigenicity of breast cancer cell lines.16
As described above, PES1 has two important roles, namely neural development and oncogenesis. These two key processes are themselves characteristics of NBs. Therefore, in the current study we investigated the relationship between PES1 and NBs. We show here that PES1 is a prognostic marker of NBs and regulates NB proliferation and apoptosis. Reduced PES1 expression is associated with NB differentiation in NB cell lines and tumorsphere models. In addition, our analysis with confocal microscopy showed that PES1 is distributed in the dense fibrillar component (DFC) in the nucleolus, and its distribution is radically changed after treatment with the DNA-damaging agent camptothecin (CPT).
Materials and Methods
Mice
The MYCN transgenic (Tg) mice17 were maintained in the animal facility at Nagoya University Graduate School of Medicine (Nagoya, Japan), where they were housed in a controlled environment and provided with standard nourishment and water. Normal ganglia and precancerous and tumor tissues from WT, hemizygous, and homozygous MYCN Tg mice were dissected and minced, and then total RNA was extracted. This study was approved by the Animal Care and Use Committee of Nagoya University Graduate School of Medicine.
Gene expression profiling
Gene expression profiling was carried out as previously described.18 Samples were analyzed with a GeneChip Mouse Genome 430 2.0 array (Affymetrix, Santa Clara, CA, USA). The data have been deposited in the Gene Expression Omnibus database of NCBI (http://www.ncbi.nlm.nih.gov/geo) under the accession number GSE43419.
Human neuroblastoma tumor profile
Kaplan–Meier analysis of the prognosis of NB patients was performed on the R2 website (http://r2.amc.nl). The dataset was from the AMC cohort study of 88 NB patients (GSE16476). This dataset was profiled on the Affymetrix HGU133 plus2.0 platform and normalized using the MASS5.0 algorithm.
Cell culture
The human neuroblastoma cell lines NB39 and TNB1 were obtained from Riken Cell Bank (Tsukuba, Japan). SH-EP and MYCN-inducible SH-EP cell line (SH-EPMYCN) cells were gifts from Dr. Schwab of the Division of Tumor Genetics, German Cancer Research Center (Heidelberg, Germany).19 They were cultured with DMEM (D5796; Sigma-Aldrich, St. Louis, MO, USA) or RPMI-1640 (R8758; Sigma-Aldrich) or a mixture of some other media following the manufacturer's instructions. Induction of MYCN was regulated by removal of tetracycline (1 μg/mL). The media were supplied with 10% heat-inactivated FBS in an incubator with humidified air at 37°C with 5% CO2. They were routinely authenticated on the basis of their viability, growth rate, and morphology by microscopic examination.
RNA interference
To deliver shRNA expression construct into NB cell lines, commercially available lentiviral shRNA vectors were used. Non-targeting shRNA (Sigma-Aldrich) and specific shRNAs for PES1 (Open Biosystems, Little Chalfont, UK) were used. The shRNA sequences are listed in Table S1.
Plasmids and their transfection
Human MYCN construct was generated by PCR using the Gateway recombination system. MYCN construct was cloned into CSII-CMV-RfA-IRES-venus plasmid. This plasmid and CSII-CMV-venus control plasmid were kindly provided by Dr. Miyoshi of Riken. For producing lentiviral particles, HEK293T cells were co-transfected with shRNA plasmids or overexpression plasmid in addition to psPAX2 (Addgene, Cambridge, MA, USA) and pMD2.G (Addgene) plasmids using FuGENE HD reagent (Promega, Fitchburg, WI, USA) according to the manufacturer's instructions. The neuroblastoma cell lines were infected in the presence of 8 μg/mL polybrene (Sigma-Aldrich).
Quantitative RT-PCR
RNA extraction from cultured cells and tumorspheres, and quantitative RT-PCR were carried out as previously described.20 Quantitative PCR analyses were carried out using an MX3000P or MX3005P real-time QPCR System (Agilent, Santa Clara, CA, USA). Quantitative PCR was carried out with ReverTra Ace (Toyobo, Osaka, Japan) and Thunderbird SYBR qPCR Mix (Toyobo). The relative expression levels were acquired according to the manufacturer's instructions. The primers used are listed in Table S2.
Immunocytochemistry
Immunocytochemistry was carried out as previously described using the following primary antibodies: rabbit anti-pescadillo (A300-902A; Bethyl Laboratories, Montgomery, TX, USA), mouse anti-pescadillo (#ab88543; Abcam, Cambridge, UK), rabbit anti-nucleolin (#ab22758; Abcam), mouse anti-phospho-histone H2AX (Ser139) (#05-636; Millipore, Billerica, MA, USA), and mouse anti-neuronal class III β-tubulin (Tuj1) (MMS-435p; Covance, Princeton, NJ, USA). Alexa Fluor 488-conjugated goat anti-rabbit IgG and Alexa Fluor 594-conjugated goat anti-mouse IgG were used as secondary antibodies. Hoechst 33258 was used to visualize nuclei.
Primary culture of tumorspheres from primary tumors of MYCN transgenic mice
The procedure was carried out as previously described.21 Briefly, dissected tumor tissue was minced and digested with 0.25% trypsin (Sigma-Aldrich). To obtain tumorspheres from the primary tumors, the cells were suspended in DMEM/F12ham (Sigma-Aldrich) plus 10 ng/mL epidermal growth factor, 15 ng/mL basic fibroblast growth factor (Peprotech, Rocky Hill, NJ, USA), 2% B27 supplement, 1% penicillin/streptomycin (PS; Gibco, Waltham, MA, USA), 15% FBS (HyClone, Waltham, MA, USA), 1% non-essential amino acid, 1% sodium pyruvate, and 55 μM β-mercaptoethanol. Cells were cultured in a non-treated Petri dish in a 37°C, 5% CO2 tissue culture incubator. To induce differentiation, spheres were collected and resuspended with medium containing DMEM/F12ham, 1% FBS, 2% B27 supplement, 1% sodium pyruvate, 103 U/mL leukemia inhibitory factor (LIF), and 1% PS. The spheres were seeded into a poly-D-lysine/laminin/fibronectin pre-coated dish. After 12 h the medium was changed to differentiation medium containing DMEM/F12 Ham, 1% FBS, 2% B27 supplement, 1% N2 supplement, 50 ng/mL nerve growth factor (NGF), 50 ng/mL neurotrophin-3 (NT3), and 1% PS.
TUNEL assay
The TUNEL assay was carried out using the APO-DIRECT kit (556381; BD Pharmingen, San Jose, CA, USA). Cell fixation and staining was carried out following the manufacturer's instructions. The cells were analyzed with BD FACSCalibur (BD Pharmingen) and BD CellQuest Pro software (BD Biosciences, San Jose, CA, USA).
Data analysis
Results are expressed as the mean ± SD. Their homoscedasticities were checked by the f-test. Statistical significance was evaluated with a two-tailed, unpaired Student's t-test.
Results
PES1 expression increased with NB tumor development in MYCN Tg mouse model
To examine the relationship between NB formation and PES1 expression, we used a MYCN Tg mouse model.17 In this model, human MYCN is overexpressed under the tyrosine hydroxylase promoter, which is active in migrating neural crest cells. At 2 weeks of age, the superior mesenteric ganglion (SMG) of WT mice and hemizygous or homozygous MYCN Tg mice are similar in size. However, compared to those of 2-week-old WT mice, the SMGs of 2-week-old homozygote MYCN Tg mice show accumulation of immature neuroblasts microscopically. These SMGs can be regarded as “precancerous” or “hyperplastic” lesions.18,20,22,23 Finally, at age 6–14 weeks, 100% (for homozygotes) or approximately 70% (hemizygotes) of Tg mice develop NB and die. Histology of tumors of both hemizygous and homozygous MYCN Tg mice are similar and equivalent to human neuroblastomas.23 We first examined the expression profiles of the SMGs of 2-week-old WT mice, the SMGs of 2-week-old homozygous MYCN Tg mice, and the terminal tumors of 6-week-old hemizygous or homozygous MYCN Tg mice (GSE43419). The precancerous lesions (2-week-old homozygous MYCN Tg mice SMG) showed higher Pes1 expression than the normal ganglia, and the terminal tumors of hemizygous or homozygous MYCN Tg mice showed even higher Pes1 expression than the precancerous ganglia or WT ganglia (Fig.1). This suggests that Pes1 expression increased as the NBs grew in the MYCN Tg mouse model.
Figure 1.

Pes1 expression in a neuroblastoma mice model. Results of a microarray analysis of the relative expression levels of Pes1 in superior mesenteric ganglia of wild-type mice (wt), 2-week homozygous MYCN transgenic (Tg) mice (precancerous lesions) and terminal tumors (hemizygous and homozygous MYCN Tg mice). hemi, hemizygous MYCN Tg mice; homo, homozygous MYCN Tg mice.
PES1 expression associated with poor prognosis of NB
We then investigated PES1 expression in human NB cases. For this analysis we used the database from an AMC cohort study of 88 NB patients (GSE16476) and the R2 genomics analysis and visualization platform. Expression of PES1 was significantly higher in the MYCN-amplified cases (P = 6.9 × 10−10; Fig.2a). MYCN amplification has been associated with rapid disease progression and unfavorable prognosis.4 As the PES1 promotor region has an E-box, the MYCN binding sequence, we examined whether MYCN induction enhances PES1 expression. MYCN induction in SH-EPMYCN,19 SY5Y and SH-EP cell lines significantly increased PES1 expression (Fig. S1). This result was consistent with Figure2(a). Tumor stage is also an important prognostic factor of NBs. Neuroblastomas were previously classified based on the International Neuroblastoma Staging System (INSS).24 Although this classification is widely accepted, in 2009, the International Neuroblastoma Risk Group proposed a new staging system.25 Both classifications include the designations 4S or MS, in addition to 4 and M, which indicate metastatic diseases. The S is short for “special,” and indeed the 4S and MS stages are unique to NBs. The 4S (or MS) cases show favorable prognosis and spontaneous regression in spite of multiple metastases of the skin, liver, or bone marrow. We investigated the PES1 expression in each of the INSS stages using the same cohort. Stage 4 cases showed higher levels of PES1 expression than stage 1–3 cases (P = 0.008; Fig.2b). In addition, compared to stage 4S cases, stage four cases also showed significantly higher PES1 expression (P = 0.016; Fig.2b). The correlation of PES1 expression with MYCN status or clinical stage raised the possibility that PES1 might be a good prognostic marker. We therefore analyzed the prognostic value of PES1 expression using Kaplan–Meier survival curves. High PES1 expression was significantly associated with poorer overall survival (P = 8.4 × 10−5 after Bonferroni correction; Fig.2c) and relapse-free survival (P = 5.4 × 10−3 after Bonferroni correction; Fig.2d). Previous reports have indicated that malignant tumors express higher PES1 levels than normal tissues or benign tumors in other types of cancers.9,14–16,26 In line with previous findings, these results indicate that PES1 expression is correlated with MYCN status and clinical stage, and that PES1 is a prognostic marker of NBs.
Figure 2.
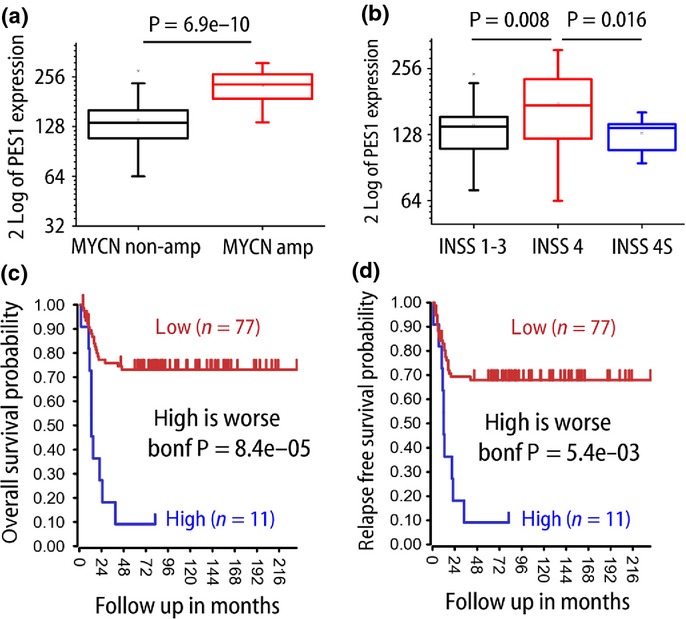
PES1 expression in human neuroblastoma cases and patient prognosis (AMC cohort, GSE16476). (a) PES1 expression in MYCN non-amplified/amplified cases (n = 72, 16 each). (b) PES1 expression in each International Neuroblastoma Staging System (INSS) stage (INSS 1–3, n = 36; INSS 4, n = 40; INSS 4S, n = 12). (c,d) Kaplan–Meier curves and log–rank test for overall survival (c) and relapse-free survival (d). Bonf, after Bonferroni correction.
PES1 knockdown suppressed NB cell growth and induced apoptosis
As NB cases with high PES1 expression are associated with unfavorable prognosis, we next investigated the role of PES1 in NB cell lines. We knocked down PES1 expression using two different shRNAs against human PES1. The shRNAs efficiently knocked down PES1 expression at both the mRNA and protein level (Fig.3a,b). Short hairpin RNAs suppressed the growth of both a MYCN-amplified cell line (NB39) and a MYCN non-amplified cell line (SH-EP) (Fig.3c,d). We next carried out a TUNEL assay to examine whether these growth suppressions were caused by apoptosis. Compared to non-targeting shRNA, shRNAs against PES1 dramatically increased the percentage of TUNEL-positive cells (Fig.3e,f). These data suggest that PES1 knockdown inhibited NB cell growth and this inhibition was caused by apoptosis.
Figure 3.
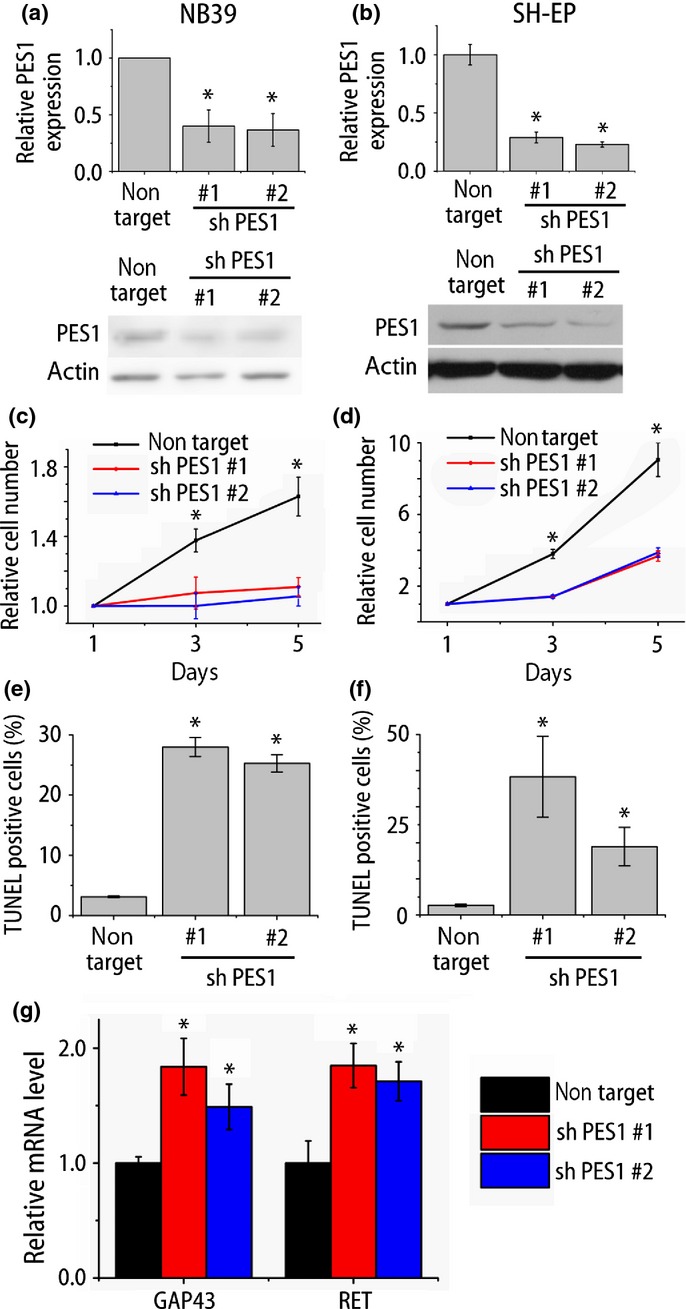
PES1 knockdown inhibited neuroblastoma (NB) cell growth and induced apoptosis. (a,b) Quantitative RT-PCR analysis and Western blot analysis of PES1 expression in NB cell lines (NB39 and SH-EP) after shRNA treatments (n = 3). (c,d) Cell proliferation analysis of NB cell lines after treatment of non-target shRNA or shRNA against PES1 (NB39, n = 4; SH-EP, n = 3). (e,f) Quantification of TUNEL-positive apoptotic cells (n = 3). (g) Quantitative RT-PCR analysis of differentiation-related gene expressions of NB39 (n = 4). *P < 0.05 non-target versus shRNA against PES1 (Student's t-test).
As already described, tumor cell differentiation is both a characteristic and an important treatment strategy of NB.3 Accordingly, we next investigated whether PES1 knockdown causes the differentiation of NB cells. Differentiated cells possess more and longer neurites and express higher levels of differentiation markers (GAP43, RET).27 Although PES1 knockdown did not induce clear increase in neurite outgrowth (data not shown), the results of quantitative PCR showed an increase in the expression of differentiation-related genes (Fig.3g). As the cell shape was not remarkably changed, we could not conclude that PES1 knockdown caused the differentiation. But this result raised the possibility that PES1 expression is associated with differentiation status.
Differentiation induction in NB cell lines decreased PES1 expression
We then examined whether the induction of differentiation can change the PES1 expression. Historically, various methods have been used to induce NB differentiation, including serum starvation and treatment with all-trans retinoic acid (ATRA).28–31 A derivative of vitamin A, ATRA is essential for development and differentiation. All-trans retinoic acid-induced differentiation of leukemic cells has been shown to dramatically improve the prognosis of patients with acute promyelocytic leukemia.32 In addition, ATRA can differentiate NB cells. We induced differentiation in NB cell lines with serum reduction (from 10% FBS in original culture to 1%) and ATRA (1 μM). This treatment increased the number and length of the neuritic extensions of NB39 and TNB1 cells (Fig.4a, Fig. S2), and these neurites were positive for Tuj1, a neuronal marker. This differentiation induction was further verified by the increased expression of a differentiation-related gene (Fig.4b, Fig. S2).33 Interestingly, the differentiation induction decreased PES1 expression (Fig.4b, Fig. S2). This result suggests the possibility that ATRA-induced differentiation modulates PES1 expression.
Figure 4.
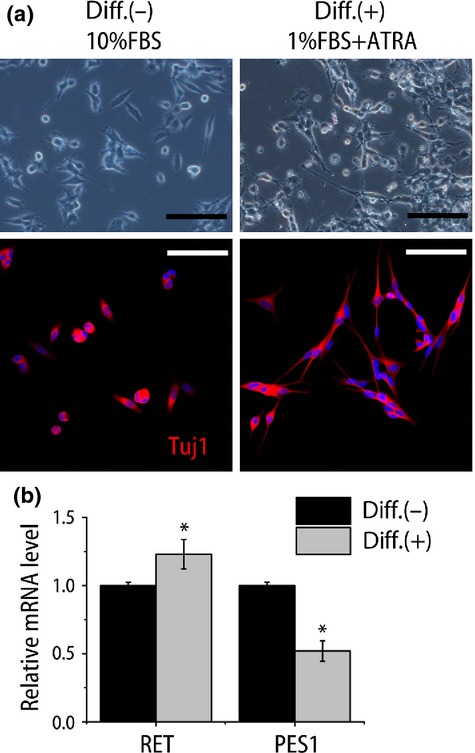
Induced differentiation attenuated PES1 expression in neuroblastoma cell line. (a) Phase contrast images (top panels) and confocal image of fluorescence immunostaining for β-tubulin (Tuj1) (red, bottom panels) of NB39 cells under normal culture conditions (10% FBS; Diff.(−)) and differentiation conditions (1% FBS + 1 μM all-trans retinoic acid [ATRA]; Diff.(+)). Hoechst 33258 was used to visualize nuclei. Scale bar = 100 μm. (b) Quantitative RT-PCR analysis of differentiation-related gene (RET) and PES1 expressions (n = 4). *P < 0.05 normal culture conditions versus differentiation conditions (Student's t-test).
Differentiation induction in mouse NB model also decreased PES1 expression
To further verify the association of PES1 with tumor cell differentiation, we tried another NB model. We used tumorspheres derived from an MYCN Tg mouse primary tumor.21 Differentiation induction with NGF, NT3, and N2 supplement caused radial neurite outgrowth (Fig.5a). Concurrently, the expressions of differentiation marker (Tuj1) increased and stemness markers (Snail and Musashi) decreased (Fig.5b). In line with the results in the NB cell lines, Pes1 expression was reduced with the differentiation induction (Fig.5b). These results suggest that Pes1 expression was negatively correlated with the differentiation conditions in two kinds of different NB models.
Figure 5.
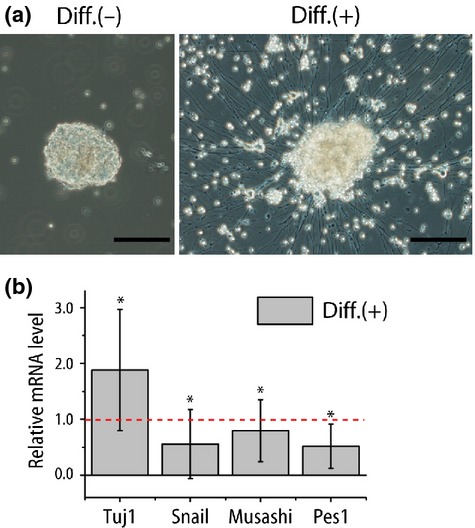
Induced differentiation decreased Pes1 expression in MYCN transgenic (Tg) mice model tumorsphere. (a) Phase contrast images of tumorspheres established from MYCN Tg mice primary tumor under normal culture conditions and differentiation conditions (NGF, NT3 addition). Scale bar = 100 μm. (b) Quantitative RT-PCR analysis of differentiation (β-tubulin, Tuj1) or stemness-related gene (Snail and Musashi) and Pes1 expressions (n = 4). Only the values of differentiation conditions are shown (normal culture condition = 1.0). The result is the average of four independently established tumorsphere samples. *P < 0.05 normal culture conditions versus differentiation conditions (Student's t-test).
PES1 localized in DFC in nucleolus and DNA damage induced redistribution
As previously reported, PES1 was localized in the nucleolus within the nucleus, where ribosome neogenesis takes place.9,11 As one of the major roles of PES1 is 32S pre-rRNA processing,10,34 this distribution is reasonable. The nucleolus consists of three distinct subnucleolar compartments called the fibrillar center, the DFC, and the granular component (GC) from inside to outside,35 and ribosome maturation also proceeds from inside to outside. A previous analysis using tagged PES1 showed its GC distribution.11 However, we found that endogenous PES1 is surrounded by nucleolin, a major nucleolar protein that is found in DFC and GC,36 with nucleolin showing “donut-like” fluorescence (Fig.6a). Considering the fact that pre-rRNA resides in the DFC, this result suggest that PES1 is mainly localized in DFC of the nucleolus, not in the GC.
Figure 6.
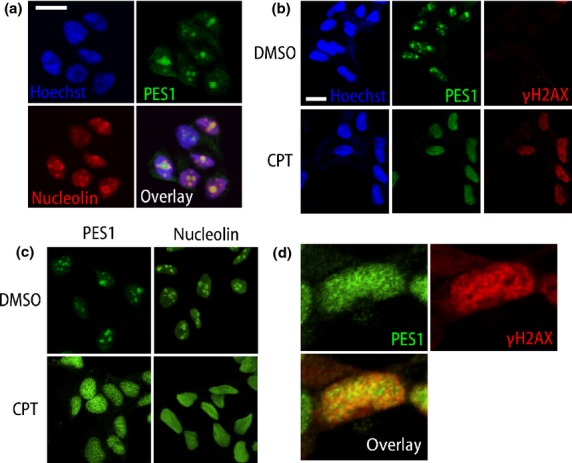
Endogenous PES1 localization and its redistribution after DNA damage. (a) Confocal images of TNB1 neuroblastoma cells immunostained for PES1 (green) and nucleolin (red) with nuclear stain (Hoechst33258, blue). PES1 is encircled by nucleolin, which shows “donut-like” staining. (b,c) Confocal images of TNB1 cells after DMSO or camptothecin (CPT; 1 μM) treatment. Scale bar = 15 μm. (d) Enlarged confocal images of TNB1 cells after CPT (1 μM) treatment. One nucleus was enlarged.
In addition to its principal role, ribosome neogenesis, the nucleolus also works as a stress sensor of DNA damage.37,38 A previous report found that PES1 knockdown induced the expression of phosphorylated H2AX (γH2AX), a DNA damage marker, and delayed the recovery from DNA damage.39 The authors indicated some association between PES1 and DNA damage response, but the relationship is still unclear. Therefore, we next investigated the dynamics of PES1 under DNA damage. Camptothecin is a topoisomerase-1 inhibitor that causes DNA damage and is used to treat cancers.37,40 In the interphase TNB1 cells, PES1 is located in the nucleolus. After treatment with CPT (1 μM) for 5 h, γH2AX was expressed strongly in the nucleus and formed γH2AX foci (Fig.6b).41 PES1, together with nucleolin, showed a radical change in distribution to a diffuse nucleoplasmic distribution (Fig.6b,c). As DNA-damaging agents are known to cause nucleolar disruption,42 this result was not surprising. The reason for this distribution change and its biological significance remains to be investigated. This result suggests that DNA damage can redistribute PES1 as well as nucleolin. PES1 expression partially, but not completely, overlapped with γH2AX (Fig.6d).
Discussion
The present study addressed the roles of PES1 in NB tumorigenesis by using a clinical NB database of mRNA expression and survival, human NB cell lines, an NB model in MYCN Tg mice, and tumorspheres derived from MYCN Tg mice. MYCN Tg mice are a reliable NB model that recapitulates the clinical features of NB, histology, and molecular changes.17,43 Our study revealed that PES1 is involved in NB cell growth, survival, and differentiation. Its high expression is associated with poor prognosis. PES1 knockdown suppresses NB tumor cell growth and induces apoptosis. In their differentiated state, the NB cell lines and tumorspheres show lower PES1 expression. In contrast, PES1 knockdown induces differentiation features. In addition, we found PES1 distribution in DFC in the nucleolus and diffuse redistribution into the nucleoplasm after CPT treatment.
Our study showed that PES1 is a prognostic marker of NBs. Neuroblastoma cases with unfavorable prognosis (MYCN amplified or INSS 4) show high PES1 expression. Previous reports indicated that cancerous tissue expressed more PES1 than normal tissue or benign tumors.9,14–16,26 However, these results were based on the results of tissue microarray or cell lines. We showed the relationship between PES1 expression and prognosis in NBs in this study for the first time. As previously shown in colon cancer cell lines,26 PES1 knockdown strikingly suppressed NB cell growth in our study.
We showed that PES1 knockdown upregulated some differentiation-related genes. To further elucidate the relationship between PES1 expression and differentiation, we used two different kinds of models to induce NB differentiation. One is ATRA treatment of NB cell lines, and the other is tumorsphere differentiation with growth factors. We previously reported a new tumorsphere culture condition,21 and found that this condition worked well in the present experimental scheme. In both models, we successfully induced differentiation as demonstrated by morphology change (neurite growth) and differentiation marker expression. Under these conditions, PES1 was downregulated. However, we could not successfully rescue these phenomena by PES1 overexpression (data not shown). These results suggest that PES1 is required but not sufficient to suppress NB cell differentiation.
We also reanalyzed PES1 distribution in the nucleoli using confocal microscopy. In our analysis, PES1 was encircled by nucleolin. Nucleolin and nucleophosmin are the two major protein components of the nucleolus. Previous reports have described multiple functions of nucleolin from rRNA splicing to ribosome assembly. Our results indicate that PES1 is mainly distributed in DFC, but not GC, whereas nucleolin is localized in DFC and GC. As DFC is an rRNA-rich lesion in nucleoli, the DFC distribution of PES1 makes sense. We also observed a dynamic change of PES1 distribution after treatment with the DNA-damaging agent CPT. The relationship between PES1 and DNA damage response is still unclear. One previous study indicated that PES1 works protectively against DNA damage and PES1 knockdown induces γH2AX expression.39 From our results and previous knowledge, we can hypothesize that PES1 knockdown causes DNA damage and accumulated DNA damage drives cells to apoptosis.
Another interesting fact is that PES1 has been known to have a BRCA1 C-terminal domain (BRCT domain).12 The BRCT domain is a phosphoprotein binding domain and an integral signaling module in the DNA damage response.44 As the role or binding partners of the BRCT domain of PES1 is not known, we investigated the binding partners of PES1 using liquid chromatography–mass spectrometry with peptide mass fingerprinting (LC-MS/MS). At this stage, NONO and SFPQ are among the list of identified proteins as binding partners (data not shown). These proteins form heterodimers and enhance DNA strand break rejoining.45,46 These results also suggested the possible close association of PES1 with DNA damage. But we need further study to elucidate the role of PES1 in DNA damage response.
In this article, we described multiple functions of the PES1 protein. The classical and best-known role of PES1 is ribosome genesis.10,11. The ribosome is an essential intracellular organelle to protein synthesis, and its dysfunction cause severe cellular disturbance. The growth inhibitions and apoptosis observed in our study (Fig.3) might be caused by this ribosomal dysfunction. In addition, as mentioned above, DNA damage caused by PES1 knockdown could also be the cause of apoptosis and growth inhibitions. Arrest of cell division is a prerequisite to enter a program of differentiation.47 Therefore, the phenotypes related to differentiation (Figs. 3–5) are closely linked to growth inhibition.
In summary, we showed that PES1 is a good prognostic marker of NBs. As a multifunctional protein, PES1 is involved in NB cell survival and differentiation. DNA damage might contribute to apoptosis regulation in NB cells.
Acknowledgments
This work was supported in part by a grant-in-aid from the National Cancer Center Research and Development Fund (22-4 to KK) and Health and Labor Sciences Research Expenses for Commission, Applied Research for Innovative Treatment of Cancer (H26-applied-general-006) from the Ministry of Health, Labor and Welfare.
Disclosure Statement
The authors have no conflicts of interest.
Supporting Information
Fig. S1. MYCN induction in MYCN single-copy cell lines.
Fig. S2. Induced differentiation in TNB1 neuroblastoma cells.
Table S1. Short hairpin RNA target sequences (PES1).
Table S2. Primer sequences for PCR experiments.
References
- Maris JM, Hogarty MD, Bagatell R, Cohn SL. Neuroblastoma. Lancet. 2007;369:2106–20. doi: 10.1016/S0140-6736(07)60983-0. [DOI] [PubMed] [Google Scholar]
- Kocak H, Ackermann S, Hero B, et al. Hox-C9 activates the intrinsic pathway of apoptosis and is associated with spontaneous regression in neuroblastoma. Cell Death Dis. 2013;4:e586. doi: 10.1038/cddis.2013.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthay KK, Villablanca JG, Seeger RC, et al. Treatment of high-risk neuroblastoma with intensive chemotherapy, radiotherapy, autologous bone marrow transplantation, and 13-cis-retinoic acid. Children's Cancer Group. N Engl J Med. 1999;341:1165–73. doi: 10.1056/NEJM199910143411601. [DOI] [PubMed] [Google Scholar]
- Maris JM, Matthay KK. Molecular biology of neuroblastoma. J Clin Oncol. 1999;17:2264–79. doi: 10.1200/JCO.1999.17.7.2264. [DOI] [PubMed] [Google Scholar]
- Guglielmi L, Cinnella C, Nardella M, et al. MYCN gene expression is required for the onset of the differentiation programme in neuroblastoma cells. Cell Death Dis. 2014;5:e1081. doi: 10.1038/cddis.2014.42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allende ML, Amsterdam A, Becker T, Kawakami K, Gaiano N, Hopkins N. Insertional mutagenesis in zebrafish identifies two novel genes, pescadillo and dead eye, essential for embryonic development. Genes Dev. 1996;10:3141–55. doi: 10.1101/gad.10.24.3141. [DOI] [PubMed] [Google Scholar]
- Simmons T, Appel B. Mutation of pescadillo disrupts oligodendrocyte formation in zebrafish. PLoS ONE. 2012;7:e32317. doi: 10.1371/journal.pone.0032317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gessert S, Maurus D, Rossner A, Kuhl M. Pescadillo is required for Xenopus laevis eye development and neural crest migration. Dev Biol. 2007;310:99–112. doi: 10.1016/j.ydbio.2007.07.037. [DOI] [PubMed] [Google Scholar]
- Kinoshita Y, Jarell AD, Flaman JM, et al. Pescadillo, a novel cell cycle regulatory protein abnormally expressed in malignant cells. J Biol Chem. 2001;276:6656–65. doi: 10.1074/jbc.M008536200. [DOI] [PubMed] [Google Scholar]
- Du YC, Stillman B. Yph1p, an ORC-interacting protein: potential links between cell proliferation control, DNA replication, and ribosome biogenesis. Cell. 2002;109:835–48. doi: 10.1016/s0092-8674(02)00773-0. [DOI] [PubMed] [Google Scholar]
- Oeffinger M, Leung A, Lamond A, Tollervey D. Yeast Pescadillo is required for multiple activities during 60S ribosomal subunit synthesis. RNA. 2002;8:626–36. doi: 10.1017/s1355838202020022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holzel M, Grimm T, Rohrmoser M, et al. The BRCT domain of mammalian Pes1 is crucial for nucleolar localization and rRNA processing. Nucleic Acids Res. 2007;35:789–800. doi: 10.1093/nar/gkl1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimm T, Holzel M, Rohrmoser M, et al. Dominant-negative Pes1 mutants inhibit ribosomal RNA processing and cell proliferation via incorporation into the PeBoW-complex. Nucleic Acids Res. 2006;34:3030–43. doi: 10.1093/nar/gkl378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber A, Hengge UR, Stricker I, et al. Protein microarrays for the detection of biomarkers in head and neck squamous cell carcinomas. Hum Pathol. 2007;38:228–38. doi: 10.1016/j.humpath.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Kim B, Bang S, Lee S, et al. Expression profiling and subtype-specific expression of stomach cancer. Cancer Res. 2003;63:8248–55. [PubMed] [Google Scholar]
- Li J, Yu L, Zhang H, et al. Down-regulation of pescadillo inhibits proliferation and tumorigenicity of breast cancer cells. Cancer Sci. 2009;100:2255–60. doi: 10.1111/j.1349-7006.2009.01325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. EMBO J. 1997;16:2985–95. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami-Tonami Y, Kishida S, Takeuchi I, et al. Inactivation of SMC2 shows a synergistic lethal response in MYCN-amplified neuroblastoma cells. Cell Cycle. 2014;13:1115–31. doi: 10.4161/cc.27983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutz W, Stohr M, Schurmann J, Wenzel A, Lohr A, Schwab M. Conditional expression of N-myc in human neuroblastoma cells increases expression of alpha-prothymosin and ornithine decarboxylase and accelerates progression into S-phase early after mitogenic stimulation of quiescent cells. Oncogene. 1996;13:803–12. [PubMed] [Google Scholar]
- Kishida S, Mu P, Miyakawa S, et al. Midkine promotes neuroblastoma through Notch2 signaling. Cancer Res. 2013;73:1318–27. doi: 10.1158/0008-5472.CAN-12-3070. [DOI] [PubMed] [Google Scholar]
- Cao D, Kishida S, Huang P, et al. A new tumorsphere culture condition restores potentials of self-renewal and metastasis of primary neuroblastoma in a mouse neuroblastoma model. PLoS ONE. 2014;9(1):e86813. doi: 10.1371/journal.pone.0086813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang P, Kishida S, Cao D, et al. The neuronal differentiation factor NeuroD1 downregulates the neuronal repellent factor Slit2 expression and promotes cell motility and tumor formation of neuroblastoma. Cancer Res. 2011;71:2938–48. doi: 10.1158/0008-5472.CAN-10-3524. [DOI] [PubMed] [Google Scholar]
- Hansford LM, Thomas WD, Keating JM, et al. Mechanisms of embryonal tumor initiation: distinct roles for MycN expression and MYCN amplification. Proc Natl Acad Sci USA. 2004;101:12664–9. doi: 10.1073/pnas.0401083101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodeur GM, Pritchard J, Berthold F, et al. Revisions of the international criteria for neuroblastoma diagnosis, staging, and response to treatment. J Clin Oncol. 1993;11:1466–77. doi: 10.1200/JCO.1993.11.8.1466. [DOI] [PubMed] [Google Scholar]
- Monclair T, Brodeur GM, Ambros PF, et al. The International Neuroblastoma Risk Group (INRG) staging system: an INRG Task Force report. J Clin Oncol. 2009;27:298–303. doi: 10.1200/JCO.2008.16.6876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Feng Q, Su Y, et al. Transcriptional regulation of PES1 expression by c-Jun in colon cancer. PLoS ONE. 2012;7:e42253. doi: 10.1371/journal.pone.0042253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima H, Bogenmann E. Expression of trkA cDNA in neuroblastomas mediates differentiation in vitro and in vivo. Mol Cell Biol. 1993;13:7447–56. doi: 10.1128/mcb.13.12.7447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seidman KJ, Barsuk JH, Johnson RF, Weyhenmeyer JA. Differentiation of NG108-15 neuroblastoma cells by serum starvation or dimethyl sulfoxide results in marked differences in angiotensin II receptor subtype expression. J Neurochem. 1996;66:1011–18. doi: 10.1046/j.1471-4159.1996.66031011.x. [DOI] [PubMed] [Google Scholar]
- Hill DP, Robertson KA. Characterization of the cholinergic neuronal differentiation of the human neuroblastoma cell line LA-N-5 after treatment with retinoic acid. Brain Res Dev Brain Res. 1997;102:53–67. doi: 10.1016/s0165-3806(97)00076-x. [DOI] [PubMed] [Google Scholar]
- Huang S, Laoukili J, Epping MT, et al. ZNF423 is critically required for retinoic acid-induced differentiation and is a marker of neuroblastoma outcome. Cancer Cell. 2009;15:328–40. doi: 10.1016/j.ccr.2009.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frumm SM, Fan ZP, Ross KN, et al. Selective HDAC1/HDAC2 inhibitors induce neuroblastoma differentiation. Chem Biol. 2013;20:713–25. doi: 10.1016/j.chembiol.2013.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenaux P, Chastang C, Chomienne C, et al. Treatment of newly diagnosed acute promyelocytic leukemia (APL) by all transretinoic acid (ATRA) combined with chemotherapy: the European experience. European APL Group. Leuk Lymphoma. 1995;16:431–7. doi: 10.3109/10428199509054430. [DOI] [PubMed] [Google Scholar]
- Celay J, Blanco I, Lazcoz P, Rotinen M, Castresana JS, Encio I. Changes in gene expression profiling of apoptotic genes in neuroblastoma cell lines upon retinoic acid treatment. PLoS ONE. 2013;8:e62771. doi: 10.1371/journal.pone.0062771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapik YR, Fernandes CJ, Lau LF, Pestov DG. Physical and functional interaction between Pes1 and Bop1 in mammalian ribosome biogenesis. Mol Cell. 2004;15(1):17–29. doi: 10.1016/j.molcel.2004.05.020. [DOI] [PubMed] [Google Scholar]
- Raska I, Shaw PJ, Cmarko D. Structure and function of the nucleolus in the spotlight. Curr Opin Cell Biol. 2006;18:325–34. doi: 10.1016/j.ceb.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Ugrinova I, Monier K, Ivaldi C, et al. Inactivation of nucleolin leads to nucleolar disruption, cell cycle arrest and defects in centrosome duplication. BMC Mol Biol. 2007;8:66. doi: 10.1186/1471-2199-8-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubbi CP, Milner J. Disruption of the nucleolus mediates stabilization of p53 in response to DNA damage and other stresses. EMBO J. 2003;22:6068–77. doi: 10.1093/emboj/cdg579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardi R, Scaglioni PP, Bergmann S, Horn HF, Vousden KH, Pandolfi PP. PML regulates p53 stability by sequestering Mdm2 to the nucleolus. Nat Cell Biol. 2004;6:665–72. doi: 10.1038/ncb1147. [DOI] [PubMed] [Google Scholar]
- Xie W, Qu L, Meng L, Liu C, Wu J, Shou C. PES1 regulates sensitivity of colorectal cancer cells to anticancer drugs. Biochem Biophys Res Commun. 2013;431:460–5. doi: 10.1016/j.bbrc.2012.12.145. [DOI] [PubMed] [Google Scholar]
- Buckwalter CA, Lin AH, Tanizawa A, Pommier YG, Cheng YC, Kaufmann SH. RNA synthesis inhibitors alter the subnuclear distribution of DNA topoisomerase I. Cancer Res. 1996;56:1674–81. [PubMed] [Google Scholar]
- Riballo E, Kuhne M, Rief N, et al. A pathway of double-strand break rejoining dependent upon ATM, Artemis, and proteins locating to gamma-H2AX foci. Mol Cell. 2004;16:715–24. doi: 10.1016/j.molcel.2004.10.029. [DOI] [PubMed] [Google Scholar]
- Boulon S, Westman BJ, Hutten S, Boisvert FM, Lamond AI. The nucleolus under stress. Mol Cell. 2010;40:216–27. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiyonari S, Kadomatsu K. Neuroblastoma models for insights into tumorigenesis and new therapies. Expert Opin Drug Discov. 2015;10(1):53–62. doi: 10.1517/17460441.2015.974544. [DOI] [PubMed] [Google Scholar]
- Leung CC, Glover JN. BRCT domains: easy as one, two, three. Cell Cycle. 2011;10:2461–70. doi: 10.4161/cc.10.15.16312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krietsch J, Caron MC, Gagne JP, Ethier C, Vignard J, Vincent M, et al. PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 2012;40:10287–301. doi: 10.1093/nar/gks798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salton M, Lerenthal Y, Wang SY, Chen DJ, Shiloh Y. Involvement of Matrin 3 and SFPQ/NONO in the DNA damage response. Cell Cycle. 2010;9:1568–76. doi: 10.4161/cc.9.8.11298. [DOI] [PubMed] [Google Scholar]
- Peunova N, Enikolopov G. Nitric oxide triggers a switch to growth arrest during differentiation of neuronal cells. Nature. 1995;375:68–73. doi: 10.1038/375068a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1. MYCN induction in MYCN single-copy cell lines.
Fig. S2. Induced differentiation in TNB1 neuroblastoma cells.
Table S1. Short hairpin RNA target sequences (PES1).
Table S2. Primer sequences for PCR experiments.


