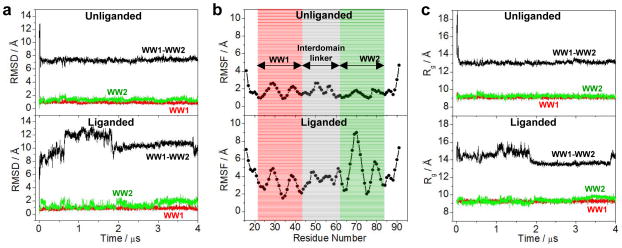
Figure 7.
Conformational dynamics as probed through MD analysis conducted on the structural model of WW1-WW2 tandem module of WWOX alone (unliganded) and in complex with p73 peptide containing the PPXY motif bound to the WW1 domain (liganded). (a) RMSD of backbone atoms (N, Cα and C) within each simulated structure relative to the initial modeled structure of unliganded (top panel) and liganded (bottom panel) WW1-WW2 tandem module as a function of simulation time. (b) RMSF of backbone atoms (N, Cα and C) averaged over the entire course of corresponding MD trajectory of unliganded (top panel) and liganded (bottom panel) WW1-WW2 tandem module as a function of residue number. Note that the red and green vertical rectangular boxes respectively demarcate the core residue boundaries (excluding the terminal loops) of WW1 and WW2 domains, while the gray vertical rectangular box denotes the residues spanning the interdomain linker. (c) Radius of gyration (Rg) of each simulated structure relative to the initial modeled structure of unliganded (top panel) and liganded (bottom panel) WW1-WW2 tandem module as a function of simulation time. Note that in (a) and (c), the overall physical parameter for each WW1-WW2 tandem module spanning residues 16–91 (black) is deconvoluted into the corresponding core regions (excluding the terminal loops) of constituent WW1 domain spanning residues 22–43 (red) and WW2 domain spanning residues 63–84 (green).