Abstract
Mutation of distal-less homeobox 3 (DLX3) is responsible for human tricho-dento-osseous syndrome (TDO) with amelogenesis imperfecta, indicating a crucial role of DLX3 in amelogenesis. However, the expression pattern of DLX3 and its specific function in amelogenesis remain largely unknown. The aim of this study was to investigate the effects of DLX3 on enamel matrix protein (EMP) genes. By immunohistochemistry assays of mouse tooth germs, stronger immunostaining of DLX3 protein was identified in ameloblasts in the secretory stage than in the pre-secretory and maturation stages, and the same pattern was found for Dlx3 mRNA using Realtime PCR. In a mouse ameloblast cell lineage, forced expression of DLX3 up-regulated the expression of the EMP genes Amelx, Enam, Klk4, and Odam, whereas knockdown of DLX3 down-regulated these four EMP genes. Further, bioinformatics, chromatin immunoprecipitation, and luciferase assays revealed that DLX3 transactivated Enam, Amelx, and Odam through direct binding to their enhancer regions. Particularly, over-expression of mutant-DLX3 (c.571_574delGGGG, responsible for TDO) inhibited the activation function of DLX3 on expression levels and promoter activities of the Enam, Amelx, and Odam genes. Together, our data show that DLX3 promotes the expression of the EMP genes Amelx, Enam, Klk4, and Odam in amelogenesis, while mutant-DLX3 disrupts this regulatory function, thus providing insights into the molecular mechanisms underlying the enamel defects of TDO disease.
Introduction
Mutation of distal-less homeobox 3 (DLX3) gene, a member of the distal-less homeodomain family (DLX1-6), is responsible for a human autosomal-dominant disease, tricho-dento-osseous syndrome (TDO; OMIM 190320) [1,2]. The most common mutation form is a 4-bp deletion (c.571_574delGGGG) in coding sequence of DLX3. TDO-affected individuals with this mutation have defects in tooth, hair, and bone, and the most penetrant phenotype features are dental findings of enamel hypoplasia and hypomaturation with taurodontism (elongation of the dental pulp chamber), suggesting a specific role of DLX3 in amelogenesis [3]. The process of amelogenesis is divided into three main phases: the pre-secretory, secretory, and maturation stages. During this process, the sequential expression and secretion of enamel matrix proteins (EMPs) are critical and considered to be co-regulated by transcriptional factors, cytokines, growth factors, and signaling molecules [4,5]. Since the hypoplastic and hypomaturation enamel defects of TDO are comparable to those caused by mutations of EMP genes such as AMELX, AMBN, ENAM, MMP20, and KLK4 [6–8], a possible link between DLX3 and EMP genes is indicated.
DLX3 is expressed in the placenta during early embryonic development, and is later found in skin and bone, as well as tissues derived from epithelial-mesenchymal interactions, including dental epithelium and mesenchyme [9,10]. As is regarded to be a transcriptional activator, DLX3 is composed of a DNA-binding homeodomain and two transactivation domains separately located in the N-terminal region or just downstream of the homeodomain [11]. Ex vivo examination of osteoblastic and keratinocyte cell lines have shown that the mutant DLX3 responsible for TDO (DLX3TDO) exerts a dominant-negative effect on the normal function of wild-type DLX3, which might lead to the abnormal phenotypes [12].
Studies have strongly suggested the pivotal role of DLX3 in controlling matrix deposition and biomineralization. In osteoprogenitor cells, DLX3 promotes the expression of bone matrix proteins such as type 1 collagen, bone sialoprotein, osteocalcin, and alkaline phosphatase [13]. In particular, chromatin immunoprecipitation (ChIP) assays have confirmed that osteocalcin is directly regulated by DLX3 [14]. During dentin development, mutant DLX3 (containing a 4-bp deletion) in transgenic mice has been reported to disrupt odontoblast cytodifferentiation and lead to odontoblast apoptosis [15]. Furthermore, ex vivo studies in odontoblasts have established a mechanistic link between DLX3 and a major dentin matrix protein, DSPP [16]. In addition, DLX3 has been shown to participate in the odontoblastic and osteogenic differentiation process of dental-derived cells [17,18].
In dental enamel, also an important mineralized tissue, DLX3 expression has been found in the pre-secretory ameloblasts of mouse molars [19]. However, the spatio-temporal expression pattern of DLX3 and its exact role in amelogenesis remain largely unknown. In the current study, we characterized the expression pattern of DLX3 in amelogenesis, and analyzed the effects of DLX3 on EMP genes in vitro. We found that in amelogenesis, DLX3 plays crucial roles through up-regulating the EMP genes Amelx, Enam, Klk4, and Odam, and that mutant-DLX3 disrupts this regulatory function.
Materials and Methods
Animals and ethics statement
This study was carried out in strict accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals (National Resource Council). All protocols were approved by the Animal Care and Use Committee of Peking University (Permit number: LA 2010–066). Neonatal ICR mice (postnatal days 1, 3, 7, and 14) in the postprandial state were anesthetized with 5 mg/100 g body weight of sodium pentobarbital, and all reasonable efforts were made to ameliorate suffering. For immunochemistry and real-time RT-PCR assay, tooth germs of the first mandibular molars were dissected under a microscope (Zeiss Stemi 2000-C, Germany).
Cell culture
The mouse ameloblast-like cell lineage, LS8, was a gift from Dr. Malcolm Snead (USC, Los Angeles, CA, USA) [20]. The LS8 cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; Gibco BRL, Gaithersburg, MD, USA) containing 10% (v/v) fetal bovine serum, penicillin (100 U/ml), and streptomycin (100 μg/ml) in a 37°C, 5% CO2 incubator, and passaged just before reaching confluence.
Plasmid construction
The pCI-neo-V5DLX3WT plasmid, expressing mouse DLX3 under a constitutive CMV promoter, was kindly provided by Dr. Maria Morasso (Developmental Skin Biology Section, NIAMS-NIH, DC, USA). The pCI-neo-V5DLX3TDO plasmid (with the TDO mutation, c.571_574delGGGG) was constructed by GeneChem Technology (Shanghai, China). Constructs of pGLEnam-E1, pGLEnam-E2, pGLAmelx-E2, pGLKlk4-E1, and pGLOdam-E2 were made by cloning the amplified PCR products of the corresponding enhancer regions (with potential DLX3 binding sites) into the pGL3-Promoter Luciferase Reporter Vector (Promega, Madison, WI, USA). Mutations of the potential DLX3 binding sites (TAATT changed to TGGTT) on pGLEnam-E1, pGLAmelx-E2, and pGLOdam-E2 were performed by TransGen Tech (Beijing, China), generating the constructs Mut pGLEnam-E1, Mut pGLAmelx-E2, and Mut pGLOdam-E2. The mutations were verified by DNA sequencing.
DLX3 immunohistochemistry
Dissected mouse tooth germs were fixed in 4% paraformaldehyde overnight at 4°C, then decalcified in 10% ethylenediaminetetraacetic acid, dehydrated in a graded series of ethanol, and embedded in paraffin. Serial sections were cut at 5 μm, and then immunohistochemistry was performed with the SP Histostain-Plus kit (ZSGB-BIO, China) based on the instructions, as described previously [21]. Briefly, the sections were deparaffinized, rehydrated, treated with citrate buffer (10 mM, pH 6.0), and microwaved. Then, 3% H2O2 was added to inactivate endogenous peroxidase, and 10% goat serum was used for blocking. After that, the sections were incubated with anti-DLX3 antibody (code: ab64953, Abcam, Cambridge, MA, USA) overnight at 4°C, or with non-relevant rabbit immunoglobulins as negative control. Goat anti-rabbit secondary antibody (ZSGB-BIO) and diaminobenzidine (Sigma, St. Louis, MO, USA) were then applied, and the slices were visualized under a light microscope (DMRB, Leica, Germany). Using the Image-Pro Plus software (version 6) (Media Cybernetics, USA), the mean optical density of the ameloblasts region [integrated optical density (IOD)/unit area] was determined, which represents the immunoreactivity of DLX3 protein within ameloblast cells.
Plasmid transfection
LS8 cells were seeded at a density of 2×104 cells per cm2 and cultured overnight. For transfection, the VigoFect reagent (Vigorous Biotech, Beijing, China) and plasmid DNA were separately diluted in opti-DMEM (Gibco BRL) for 5 min, mixed at room temperature for 15 min, and then added to the cultures following the manufacturer’s instructions. After 6 h of transfection, the medium was replaced. Cells were harvested and analyzed at 36 or 48 h post-transfection.
RNA interference
Three different nucleotides (20–23 nt, named Dlx3 siRNA #1, #2, #3) targeting mouse Dlx3 mRNA (NM_005220) were produced by Sigma and tested for silencing. A nonspecific siRNA (NS siRNA; Sigma) was used as control. LS8 cells were seeded and cultured until sub-confluence. Dlx3 siRNA or NS siRNA was diluted in DMEM to a final concentration of 50 nM, combined with the Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA), incubated at room temperature for 20 min, and then added to the cultures. The medium was replaced 8 h later, and total RNA and protein were extracted at 36 or 48 h post-transfection for further analysis. The sequences of the siRNAs were as follows: Dlx3 siRNA (#1), GUCACUGACCUGGGCUAUUdTdT, AAUAGCCCAGGUCAGUGACdTdT; Dlx3 siRNA (#2), CGAACGAUCUACUCCAGCUdTdT, AGCUGGAGUAGAUCGUUCGdTdT; Dlx3 siRNA (#3), GUGACUCCAUGG CCUGCAAdTdT, UUGCAGGCCAUGGAGUCACdTdT; NS siRNA, UUCUCCGAACGUGUCACGUTT, ACGUGACACGUUCGGAGAATT.
Western blot
LS8 cells were washed twice with PBS, and then lysed in modified radioimmunoprecipitation assay lysis buffer (Beyotime, Beijing, China) containing 1 mM phenylmethylsulfonyl fluoride. Protein concentration was quantified with the bicinchoninic acid protein assay (Pierce, Rockford, IL, USA). Aliquots of 50 μg protein extract per sample were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to nitrocellulose membranes. The membranes were blocked with 5% skimmed milk for 1 h and then incubated overnight with specific primary antibodies at 4°C. The following antibodies were used: anti-DLX3 (code: sc-18143, Santa Cruz, CA, USA), anti-amelogenin (kindly provided by Dr. DenBesten, University of California, USA), anti-enamelin (prepared as previously described) [22], and anti-GAPDH (code: TA-08, ZSGB-BIO). The membranes were then incubated with secondary sheep anti-mouse or sheep anti-rabbit horseradish peroxidase-conjugated linked antibodies (Santa Cruz) for 1 h. The protein signals were visualized with Chemiluminescent Substrate (Millipore, MA, USA) and exposed by the ChemiDoc XRS System (BioRad, CA, USA). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) served as an internal control. The densitometric analysis was performed on grayscale images from 3 independent experiments with Multi-gauge software (Fuji, Japan).
Real-time RT-PCR
Total RNA was isolated from LS8 cell cultures or mouse tooth germs using Trizol reagent (Vigorous Biotech), according to the manufacturer’s instructions. cDNA synthesis was carried out in a 25 μl reaction mixture containing 2 μg total RNA, 400 mM reverse transcription primers, 4 U/μl M-MLV, 1 U/μl RNAsin, and 0.4 mM dNTP mix, using M-MLV reverse transcriptase (Promega). The amplification reaction was carried out in an ABI 7300 Real-Time PCR System (Applied Biosystems, CA, USA) with SYBR Green Master Mix (Toyobo, Osaka, Japan) and the appropriate primers (Table 1). The annealing temperature was 60°C. Transcription levels were normalized against GAPDH, and each value is the average of three independent experiments.
Table 1. RT-PCR primers.
| Gene | Primer sequence | Species | GenBank accession number | PCR product size (bp) |
|---|---|---|---|---|
| Enam | S 5’-TGGCAATGGACTTTACCCCTATC-3’ | Mouse | NM_017468.3 | 273 |
| AS 5’-GCATCAGGCACAGTTGAGTTTGTAG-3’ | ||||
| Amelx | S 5’-TGAGGTGCTTACCCCTTTGAAGTG-3’ | Mouse | NM_009666.3 | 216 |
| AS 5’-GGAACTGGCATCATTGGTTGC-3’ | ||||
| Ambn | S 5’-TTGAGCCTTGAGACAATGAGAC-3’ | Mouse | NM_009664.1 | 114 |
| AS 5-’AAGTCCGTGCAACCATAAACTAT-3’ | ||||
| Tuft1 | S 5’-CGGAACTGGTGTACCTTGGTG-3’ | Mouse | NM_011656.2 | 152 |
| AS 5’-GGCATCATGGCATAGGTCTTC-3’ | ||||
| Mmp20 | S 5’-TACGAAGTGGCTGAACGAG-3’ | Mouse | NM_013903.2 | 114 |
| AS 5’-TGGGAACCCGAAGTCATA-3’ | ||||
| Klk4 | S 5’-TTGCAAACGATCTCATGCTC-3’ | Mouse | NM_019928.1 | 228 |
| AS 5’-TGAGGTGGTACACAGGGTCA-3’ | ||||
| Odam | S 5’-GTCACATCCTCACCACAGCA-3’ | Mouse | NM_027128.2 | 160 |
| AS 5’-GAGTTTCTGGAGCTGTGCCT-3’ | ||||
| Amtn | S 5’-GGACCACTGAATGGACAGCA-3’ | Mouse | NM_027793.1 | 191 |
| AS 5’-TCTGGTTTAGTGCCTGCCTG-3’ | ||||
| DLX3 | S 5’-AGCCCAGTATCTGGCCTTG-3’ | Mouse | NM_010055.3 | 133 |
| AS 5’-CGGCACCTCCCCATTCTTA-3’ | ||||
| Gapdh | S 5’- CCAGCCTCGTCCCGTAGACA-3’ | Mouse | NM_008084.2 | 189 |
| AS 5’- CCGTTGAATTTGCCGTGAGT-3’ |
As, antisense; S, sense.
Chromatin immunoprecipitation assay
LS8 cells were transfected with pCI-neo-V5DLX3WT for 48 h. Cells were cross-linked with 1% (v/v) formaldehyde at 37°C for 10 min. DNA was sheared by sonication and immunoprecipitated with nonspecific IgG or anti-DLX3 antibody (code: ab66390, Abcam) for 12 h at 4°C. Immune complexes were incubated with Protein A/G-Sepharose CL-4B (Amersham Biosciences, Uppsala, Sweden) for 2 h at 4°C. Protein-DNA cross-linking was reversed by overnight incubation at 65°C. The precipitated DNA was amplified by real-time PCR for fragments of the specific enhancer region. PCR products were separated onto a 2% agarose gel, visualized, and analyzed with a GelDoc-It TS Imaging System (UVP, Upland, CA, USA). The PCR primers used for chromatin immunoprecipitation (ChIP) assay are listed in Table 2.
Table 2. Primers for ChIP assay.
| Fragments | Predicted binding site | Primer sequences (5’-3’) | Product size (bp) |
|---|---|---|---|
| Enam-E1 | -4842/-4824 bp | S 5’-AAGAATGTATCAGTGGTTGG-3’ | 183 |
| AS 5’-GTTAAGCCTCAGTTTCCTCA-3’ | |||
| Enam-E2 | -4650/-4632 bp | S 5’-TGAGGAAACTGAGGCTTAAC-3’ | 123 |
| AS 5’-TATTTATGGTGTCTTCGGAT-3’ | |||
| Amelx-E1 | -5278/-5260 bp | S 5’-TCCATGGGGACATTGCATTT-3’ | 166 |
| AS 5’-ACACCTCAAATCTCAACCTTTCT-3’ | |||
| Amelx-E2 | -1146/-1128 bp | S 5’-TCTTTGTGCCATCTACACCA-3’ | 160 |
| AS 5’-CAAATCTGGCTCCCAAAAGGC-3’ | |||
| Klk4-E1 | -3711/-3693 bp | S 5’-AGCTACATCCCTCCAGCTTCA-3’ | 197 |
| AS 5’-ACAGTCTTCCCGACATGCTTC-3’ | |||
| Odam-E1 | -4492/-4474 bp | S 5’-TCTGTGAGCCTCTTGGTGGAT-3’ | 180 |
| AS 5’-CGTTCATTCACCAGCACAAAAC-3’ | |||
| Odam-E2 | -1769/-1747 bp | S 5’-AGGGATTCCATTTGCTGCAC-3’ | 193 |
| AS 5’-AGGATCACAAGTATTCTGATGAAA-3’ |
As, antisense; S, sense.
Luciferase assays
LS8 cells were seeded into 24-well plates at 1×105 per well, and cultivated until 60% confluence. Constructs of pGL3-promoter, pGLEnam-E1, pGLEnam-E2, pGLAmelx-E2, pGLKlk4-E1, pGLOdam-E2, or Mut pGLEnam-E1, Mut pGLAmelx-E2, and Mut pGLOdam-E2 were separately co-transfected into LS8 cells with pCI-neo-V5DLX3WT, pCI-neo-V5DLX3TDO, or pCI-neo, using VigoFect reagent according to the manufacturer’s instructions. The total amount of DNA per well was kept constant using pcDNA3.1 plasmid. Luciferase activity was measured by a dual luciferase assay system (Vigorous) and normalized to Renilla luciferase activity. All experiments were performed in triplicate, and each was repeated three times.
Statistical analysis
The data are presented as mean ± standard deviation (SD). Comparisons between two groups were performed on GraphPrism5 statistical software (GraphPad Software, USA), using Student’s t-test or one-way analysis of variance. A significant difference was noted when P <0.05.
Results
Spatial and temporal expression of Dlx3 during mouse amelogenesis
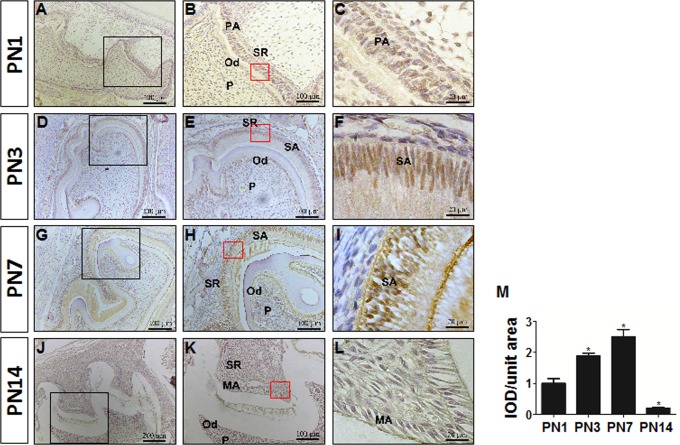
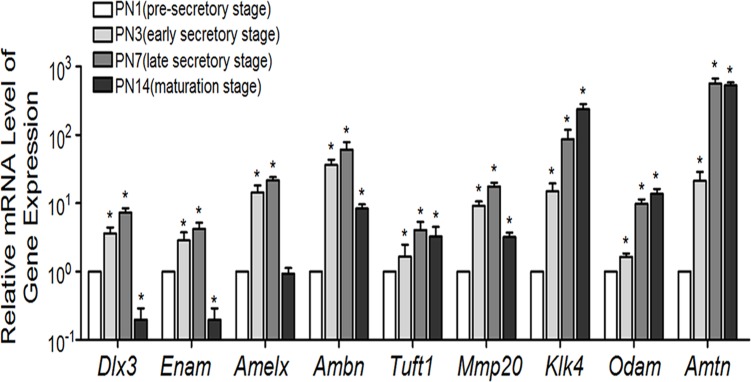
Immunostaining for DLX3 protein in mouse tooth germs of the mandibular first molar showed positive staining localized in the nuclei (Fig. 1A-L). The immunoreactivity of DLX3 staining was then quantified using Image-pro plus software (Fig. 1M). On postnatal day 1 (PN1), the enamel matrix had not been secreted in most area (except for the cusp region), indicating the pre-secretory stage of amelogenesis (Fig. 1A-C). DLX3 was weakly expressed in pre-secretory ameloblasts. At PN3, the enamel matrix was partially deposited and the cells indicated were entering the early secretory stage (Fig. 1D-F). Staining of DLX3 in early secretory ameloblasts was stronger than in pre-secretory ameloblasts. On PN7 sections, which represented the late secretory stage, DLX3 staining was even stronger in late than in early secretory ameloblasts (Fig. 1G-I). However, the staining of DLX3 significantly decreased and was hardly detectable in maturation-stage ameloblasts of PN14 mice (Fig. 1J-L). DLX3 staining was also observed in odontoblasts and dental pulp cells, but seldom in the stellate reticulum layer. Consistent with the expression pattern of DLX3 protein, the expression of Dlx3 mRNA was elevated at the early secretory stage (PN3), further increased at late secretory stage (PN7), and then decreased at the maturation stage (PN14) (Fig. 2). The expression of all analyzed EMP genes Enam, Amelx, Ambn, Tuft1, Mmp20, Klk4, Odam, and Amtn was elevated at PN3, corresponding to the elevation of Dlx3 expression (Fig. 2).
Fig 1. Immunostaining of DLX3 protein at different stages of mouse amelogenesis.
(A-L) Immunostaining of DLX3 in sections of mouse molar germs at postnatal days 1 (A-C), 3 (D-F), 7 (G-I), and 14 (J-L). Representative figures from three independent experiments are shown. (M) The immunoreactivity of DLX3 protein within ameloblast cells was expressed as the mean optical density of the ameloblasts [integrated optical density (IOD)/unit area]. Each bar represents mean ± SD. *P <0.05 vs. the control (PN1 group). A, D, G, and J, original magnification ×100; B, E, H, and K, high-power magnification of the boxed areas in the low-magnification images, original magnification ×200; C, F, I, and L, high-power magnification of ameloblasts from the area inside the boxes in B, E, H, and K, original magnification ×1000. PA, pre-secretory ameloblasts; SA, secretory ameloblasts; MA, maturation ameloblasts; Od, odontoblast; P, dental pulp; SR, stellate reticulum layer. PN, postnatal day.
Fig 2. Expression patterns of Dlx3 and EMP genes at the mRNA level during amelogenesis.
Real-time RT-PCR was used to analyze the expression levels of Dlx3 and EMP genes using RNA extracted from tooth germs at the indicated stages. The expression level at PN1 was set at 1, and the fold-changes at other stages were calculated relative to PN1. The data represent three independent experiments, and are shown as mean ± SD. *P <0.05 vs. the control (PN1 group). Enam, Enamelin; Amelx, Amelogenin; Ambn, ameloblastin; Tuft, Tuftelin-1; Mmp20, Matrix metalloproteinase 20; Klk4, kallikrein 4; Odam, Odontogenic ameloblast-associated protein; Amtn, Amelotin.
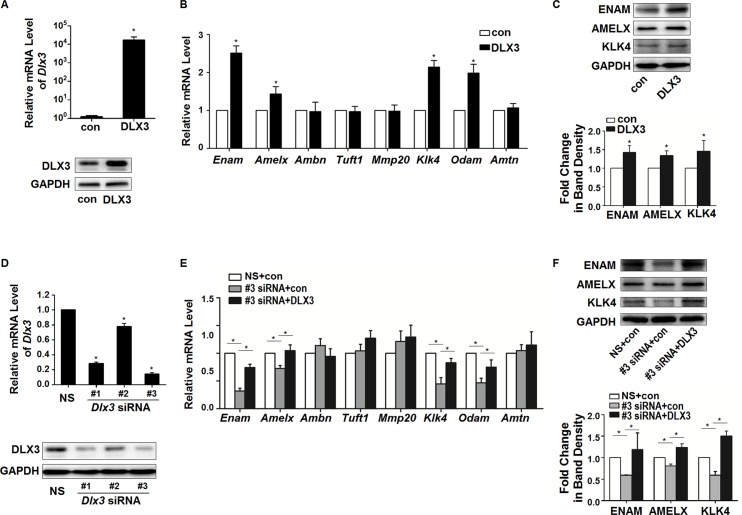
Up- and down-regulation of enamel matrix protein genes Enam, Amelx, Klk4, and Odam by DLX3-overexpression and knockdown
After 24 or 36 h of DLX3WT plasmid transfection, the over-expression efficiency of DLX3 mRNA and protein in LS8 cells was confirmed (Fig. 3A). With Dlx3-overexpression, 4 EMP genes, Enam, Amelx, Klk4, and Odam, were up-regulated by DLX3 at the mRNA level (Fig. 3B), compared with control. Western blot showed the corresponding up-regulation of ENAM, AMELX, and KLK4 proteins (Fig. 3C). Since there is no commercially-available antibody against mouse ODAM protein, it was not analyzed in this study.
Fig 3. Regulation of Enam, Amelx, Klk4, and Odam by DLX3-overexpression and knockdown.
(A) Over-expression of DLX3 was determined by real-time RT-PCR and western blot. Values are mean ± SD of the data from three independent experiments. *P <0.05 vs. control (con) group. (B) After 36 h of transfection, the expression of EMP genes was assessed. The mRNA expression of Enam, Amelx, Klk4, and Odam were significantly up-regulated. Values are presented as mean ± SD. *P <0.05 vs. the control (con) group. (C) After 48 h of transfection, elevated protein expression of ENAM, AMELX, and KLK4 were detected by western blot. Upper panel: western blot bands of ENAM, AMELX, and KLK4 (representative of three independent experiments). Lower panel: densitometric analysis of images of 3 independent experiments. *P <0.05 vs. the control (con) group. (D) Expression levels of DLX3 were determined after transfection with 3 independent Dlx3-specific small interfering RNAs (Dlx3 siRNA #1, #2, #3), or a nonspecific siRNA, NS siRNA. Values are from three independent experiments, and shown as mean ± SD. *P <0.05 vs. NS group. (E) After DLX3 silencing (using Dlx3 siRNA #3), the expression of EMP genes was assessed. The expression of Enam, Amelx, Klk4, and Odam was significantly down-regulated, and then successfully rescued by transfection of plasmid pCI-neo-V5DLX3WT. Values are mean ± SD of data from three independent experiments. *P <0.05. (F) At the protein level, expression of ENAM, AMELX, and KLK4 were also down-regulated by DLX3-knockdown and then rescued by pCI-neo-V5DLX3WT transfection, as analyzed by western blot. Upper panel: western blot bands of ENAM, AMELX, and KLK4 (representative of three independent experiments). Lower panel: densitometric analysis of images of 3 independent experiments. *P <0.05.
Expression of DLX3 in LS8 cells was successfully knocked down by siRNA fragments specific for DLX3, particularly by Dlx3 siRNA #1 and #3 (Fig. 3D). The most effective #3 Dlx3 siRNA were then used for the subsequent knockdown experiments. After DLX3 silencing, Enam, Amelx, Klk4, and Odam were down-regulated compared with the NS group, consistent with the up-regulation effects of DLX3 over-expression (Fig. 3E). Western blot analysis revealed that the protein levels of ENAM, AMELX, and KLK4 were also decreased after DLX3 knockdown (Fig. 3F). The down-regulated expression of Dlx3 and its targets genes, Enam, Amelx, Klk4 and Odam, mediated by Dlx3 siRNA, was then successfully rescued by transfection of plasmid pCI-neo-V5DLX3WT, as analyzed by qPCR and western blot, excluding the possibility of siRNA off-target effects (Fig. 3E and F).
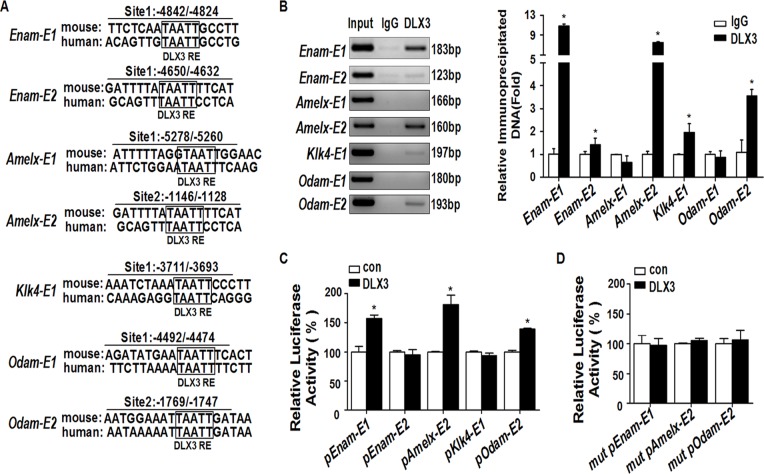
DLX3 directly transactivates the expression of Enam, Amelx, and Odam through binding to the enhancer regions
For Enam, Amelx, Klk4, and Odam, bioinformatic analysis was performed separately on 6000 bp 5’-flanking regions upstream of the translation start sites (defined as +1 bp) using MatInspector software. Conserved DLX3 potential binding sites were predicted on the enhancer regions of these 4 EMP genes, and the locations are named as follows: Enam-E1 (representing the first potential DLX3 binding site on the Enam enhancer), -4842/-4824 bp; Enam-E2, -4650/-4632 bp; Amelx-E1, -5278/-5260 bp; Amelx-E2, -1146/-1128 bp; Klk4-E1, -3711/-3693 bp; Odam-E1, -4492/-4474 bp; and Odam-E2, -1769/-1747 bp (Fig. 4A). Further ChIP assays confirmed the recruitment of DLX3 onto the predicted binding sites of Enam-E1, Enam-E2, Amelx-E2, Klk4-E1, and Odam-E2 (Fig. 4B). The primers for the distal region outside the 6000 bp 5’-flanking region of Enam, Amelx, Klk4, and Odam were designed and used as negative controls (S1 Table and S1 Fig.).
Fig 4. DLX3 directly transactivates the expression of Enam, Amelx, and Odam by binding to the enhancer regions.
(A) Bioinformatic analysis was performed on 6000 bp 5’-flanking regions of Enam, Amelx, Klk4, and Odam. 1 or 2 conserved DLX3 response elements were separately found on the four genes (translation start site defined as +1 bp). RE, response element. (B) ChIP assays determined whether DLX3 was recruited to the predicted enhancer sites. Left panel: gel images of PCR products, representative of three independent experiments. Right panel: statistics of the PCR results. Values are from three independent experiments, and shown as mean ± SD. *P <0.05 vs. IgG group. (C) Luciferase assays were performed to evaluate the impact of DLX3 on the transcriptional activity of each constructed luciferase reporter, which contained enhancer sequences of specific potential DLX3 response elements. Data are compared with the control group (con), and presented as mean% ± SD. *P <0.05. pEnam-E1 represents pGLEnam-E1, etc. (D) Mutations of DLX3 binding sites reduced the activation effect of DLX3 on the transcriptional activity of the pGLEnam-E1, pGLAmelx-E2, and pGLOdam-E2 reporters. The data represent the mean% ± SD of three independent experiments, each experiment performed in triplicate. *P <0.05 vs. the control (con) group. mut pEnam-E1 represents Mut pGLEnam-E1, etc.
To characterize the functional significance of this DLX3 recruitment, luciferase assays were performed with pGL3-promoter luciferase reporters covering the binding site and the surrounding sequence of ~200 bp. The results showed the transcriptional activation of pGLEnam-E1, pGLAmelx-E2, and pGLOdam-E2 by DLX3 recruitment, but not pGLEnam-E2 and pGLKlk4-E1, compared with control (Fig. 4C). This DLX3-dependent activation of Enam, Amelx, or Odam was also dose-dependent (S2 Fig.). These activation effects were disrupted by mutations of the DLX3 binding sites on pGLEnam-E1, pGLAmelx-E2, and pGLOdam-E2 (Fig. 4D).
TDO mutation of DLX3 inhibits the activation effects of wild-type DLX3 on EMP genes
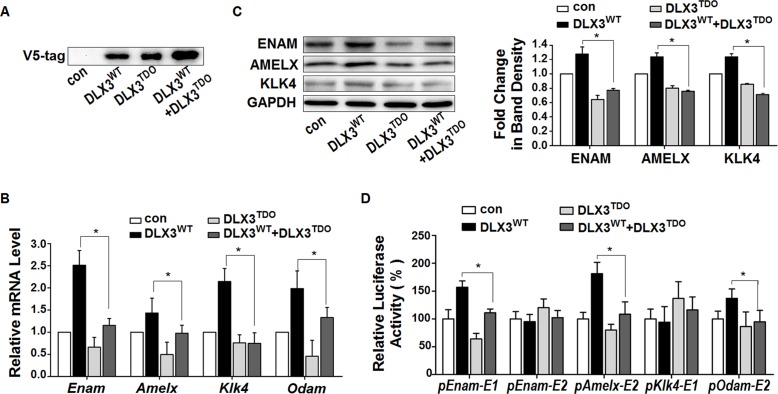
In osteoblast and keratinocyte cell lines, the mutated DLX3TDO is known to have a dominant-negative effect on the transcriptional function of DLX3WT [17,18]. To determine the influence of DLX3TDO on the activating action of DLX3WT in ameloblast-like cells, the V5DLX3TDO plasmid was transfected alone or co-transfected with the V5DLX3WT plasmid at a ratio of 1:1 into LS8 cells. The over-expression efficiency of V5DLX3WT or/and V5DLX3TDO was confirmed by western blot using an antibody against V5-tag (Fig. 5A).
Fig 5. TDO mutation of DLX3 inhibits the activation of EMP genes by wild-type DLX3.
(A) The plasmids pCI-neo (control empty vector, con), V5DLX3WT, and V5DLX3TDO were transfected into LS8 cells. Equal amounts of V5DLX3WT or V5DLX3TDO plasmid were added to each group, and the total amounts of plasmid in the groups were kept constant using pCI-neo. Over-expression of V5DLX3WT or/and V5DLX3TDO were analyzed by western blot using antibody against V5-tag. A representative figure from three independent experiments is shown. (B) After over-expression, the mRNA expression levels of Enam, Amelx, Klk4, and Odam were evaluated. Values were obtained from three independent experiments, and are presented as mean ± SD. *P <0.05 between the DLX3WT group and the DLX3WT+DLX3TDO group. (C) Left panel: western blot for the protein expression levels of ENAM, AMELX, and KLK4 after transfection (representative of three independent experiments). Right panel: densitometric analysis of images of 3 independent experiments. *P <0.05 between the DLX3WT group and the DLX3WT+DLX3TDO group. (D) After co-transfection with pCI-neo (empty control vector), V5DLX3WT, V5DLX3TDO, or both V5DLX3WT and V5DLX3TDO plasmids, the relative transcriptional activity of the 5 reporter constructs containing potential DLX3 response elements were analyzed by luciferase assays. Data were compared with the control group (con), and are presented as mean% ± SD. *P <0.05 between the DLX3WT group and the DLX3WT+DLX3TDO group. pEnam-E1 represents pGLEnam-E1, etc.
In the presence of both DLX3WT and DLX3TDO, the mRNA levels of Enam, Amelx, Klk4, and Odam were significantly reduced compared with DLX3WT alone (Fig. 5B). Correspondingly, the protein levels of ENAM, AMELX, and KLK4 were down-regulated in the DLX3WT+DLX3TDO group compared with the DLX3WT group (Fig. 5C). Further luciferase assays revealed that the transcriptional activity of Enam-E1, Amelx-E2, and Odam-E2 in the presence of both DLX3WT and DLX3TDO were decreased compared with the DLX3WT group (Fig. 5D). These results indicated that in LS8 cells, DLX3TDO inhibits the transcriptional activation of EMP genes by DLX3WT.
Discussion
In the current study, we investigated the specific role of the transcriptional factor DLX3, and the expression patterns of Dlx3 mRNA and protein during amelogenesis. Three EMP genes (Enam, Amelx, and Odam) were confirmed to be new direct targets of DLX3. The DLX3 mutant responsible for TDO interfered with the normal activation function of wild-type DLX3 in an ameloblast cell line, which partially explains the dental enamel defects in individuals with TDO.
Previous studies showed that DLX3 is expressed in mouse ameloblasts [9], yet the spatio-temporal expression of DLX3 in amelogenesis has not been reported. Our immunochemistry results revealed that DLX3 staining was strong in the nuclei of ameloblasts during secretory stages (PN3 and PN7), yet were relatively weak in the pre-secretory and maturation stages, consistent with the mRNA pattern. This expression pattern is in accord with the expression of EMP genes, suggesting us an association between DLX3 and EMP genes. Subsequent gain-of-function and loss-of-function studies identified that DLX3 promotes the expression of Enam, Amelx, Klk4, and Odam in ameloblast cell line, while mutant DLX3 inhibits this promoting function. Among these 4 EMP genes, Amelx and Enam are both associated with matrix apposition and mineralization, while Odam and Klk4 are generally thought to participate in matrix mineralization [23]. Combined with the hypoplasia and hypomaturation types of amelogenesis imperfecta detected in TDO patients [3], we suggested that mutant DLX3 disrupts both the apposition and mineralization processes of amelogenesis through inhibiting the activation function of wild-type DLX3 on Enam, Amelx, Klk4, and Odam. DLX3 staining was also observed in odontoblasts, consistent with the regulation of Dspp (dentin sialophosphoprotein), one of the main dentin matrix protein genes, by DLX3 [16].
AMELX is the most abundant EMP expressed by secretory and early maturation ameloblasts, comprising >90% of the unmineralized enamel matrix, and is thought to form an organic scaffold that is essential for regulation of crystallite growth [24]. In humans, variety of mutations in AMELX gene are associated with enamel hypoplasia and/or hypomaturation (OMIM 301200) [25]. Recently, in vitro studies have demonstrated that another DLX member, DLX2, transactivates Amelx expression, yet the expression of DLX2 is very weak at the secretory stage and is elevated at the maturation stage [26]. Since our results showed the expression of DLX3 is high at the secretory stage and decreased at the maturation stage, it seems that the activation of Amelx expression at the secretory stage is mainly controlled by DLX3, and switches to DLX2 at the maturation stage.
ENAM, the largest EMP, is also expressed at the secretory and early-maturation stages, and is considered to participate in crystallite growth and elongation [27]. Clinically, multiple ENAM mutations cause smooth hypoplastic and local hypoplastic AI (OMIM 104500) with a grossly reduced enamel volume [28]. Transgenic studies found that the -5200~-3900 bp region of the Enam enhancer is related to its tissue-specific regulation [29], consistent with our finding that DLX3 transactivated Enam expression through binding to its -4842/-4824 bp enhancer (just between the -5200~-3900 bp region). This indicates DLX3 participates in the tissue-specific expression of Enam in ameloblasts.
ODAM is a structural protein considered to be crucial in enamel mineralization [30], no mutation of ODAM gene has been associated with AI till now. Our results showed that the expression of Odam remained at a high level in the late-maturation stage, yet Dlx3 expression was very weak at this time. At different stages of osteoblast differentiation, the regulation of RUNX2 and osteocalcin are co-regulated by the interaction and interplay between DLX3, DLX5, and MSX2 [14,31]. Thus, the regulation of Odam in amelogenesis at the late-maturation stage could be controlled by other transcription factors together with DLX3, for example, other DLX members, or MSX family members (another homeodomain family) [32].
KLK4 is one of the extracellular proteases that cleave structural proteins and function in enamel maturation and mineralization [33]. Mutations in KLK4 gene are associated with pigmented hypomaturation AI (OMIM 204700) and the enamel is not fully mineralized [34]. Though the expression level of Klk4 was affected by DLX3 over-expression or knockdown, the enhancer of Klk4 was not activated by DLX3, suggesting Klk4 maybe an indirect target of DLX3. The factor(s) mediating between DLX3 and Klk4 remains to be clarified in further studies.
In addition to EMPs, interactions between epithelial ameloblasts and mesenchymal odontoblasts also affect the amelogenesis process. Disorders of odontoblast function inhibit the development of dental enamel, and vice versa [35,36]. In transgenic mice expressing DLX3TDO under a Col1A1 promoter, DLX3TDO that expressed in odontoblasts only (not ameloblasts) does not influence the enamel phenotype [15]. This suggests that the enamel disorders in TDO are caused by the specific influence of DLX3TDO on amelogenesis, in line with our results that DLX3TDO inhibited the activation of EMP genes by DLX3WT. Due to placental defects, DLX3-knockout mice die early, on embryonic day 9.5 [37]. To in vivo confirm the function of DLX3 in amelogenesis, conditional DLX3-knockout or DLX3-mutant models are being constructed for further studies.
In conclusion, we found that DLX3 up-regulates the EMP genes Amelx, Enam, Odam, and Klk4 by directly binding to their enhancer regions or indirect mechanisms, while mutant-DLX3 (responsible for TDO) disrupts this regulatory function. Our results elucidate the specific function of DLX3 in amelogenesis and provide insights into the molecular mechanisms underlying the enamel defects in TDO disease.
Supporting Information
(XLS)
(DOC)
The data represent mean ± SD of three independent experiments, each performed in triplicate.
(TIF)
The data represent mean ± SD of three independent experiments, each performed in triplicate.
(PNG)
As, antisense; S, sense.
(PNG)
Acknowledgments
We thank Dr. Malcolm Snead and Dr. Maria Morasso for the gift of LS8 cells and pCI-neo-V5DLX3WT plasmid, respectively.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This study was supported by National Natural Science Foundation of China (NSFC 81300839) (http://www.nsfc.gov.cn/); Specialized Research Fund for the Doctoral Program of Higher Education (20130001120109) (http://www.cutech.edu.cn/cn/index.htm). The funders above had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Price JA, Bowden DW, Wright JT, Pettenati MJ, Hart TC. Identification of a mutation in DLX3 associated with tricho-dento-osseous (TDO) syndrome. Hum Mol Genet. 1998;7: 563–569. [DOI] [PubMed] [Google Scholar]
- 2. Nieminen P, Lukinmaa PL, Alapulli H, Methuen M, Suojarvi T, Kivirikko S, et al. DLX3 homeodomain mutations cause tricho-dento-osseous syndrome with novel phenotypes. Cells Tissues Organs. 2011;194: 49–59. 10.1159/000322561 [DOI] [PubMed] [Google Scholar]
- 3. Al-Batayneh OB. Tricho-dento-osseous syndrome: diagnosis and dental management. Int J Dent. 2012;2012: 514692 10.1155/2012/514692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Miletich I, Sharpe PT. Normal and abnormal dental development. Hum Mol Genet. 2003;12 Spec No 1: R69–73. [DOI] [PubMed] [Google Scholar]
- 5. Bei M. Molecular genetics of ameloblast cell lineage. J Exp Zool B Mol Dev Evol. 2009;312B: 437–444. 10.1002/jez.b.21261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu JC, Chun YH, Al Hazzazzi T, Simmer JP. Enamel formation and amelogenesis imperfecta. Cells Tissues Organs. 2007;186: 78–85. [DOI] [PubMed] [Google Scholar]
- 7. Wright JT, Torain M, Long K, Seow K, Crawford P, Aldred MJ, et al. Amelogenesis imperfecta: genotype-phenotype studies in 71 families. Cells Tissues Organs. 2011;194: 279–283. 10.1159/000324339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poulter JA, Murillo G, Brookes SJ, Smith CE, Parry DA, Silva S, et al. Deletion of ameloblastin exon 6 is associated with amelogenesis imperfecta. Hum Mol Genet. 2014;23(20): 5317–5324 10.1093/hmg/ddu247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao Z, Stock D, Buchanan A, Weiss K. Expression of Dlx genes during the development of the murine dentition. Dev Genes Evol. 2000;210: 270–275. [DOI] [PubMed] [Google Scholar]
- 10. Duverger O, Morasso MI. Role of homeobox genes in the patterning, specification, and differentiation of ectodermal appendages in mammals. J Cell Physiol. 2008;216: 337–346. 10.1002/jcp.21491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Beanan MJ, Sargent TD. Regulation and function of Dlx3 in vertebrate development. Dev Dyn. 2000;218: 545–553. [DOI] [PubMed] [Google Scholar]
- 12. Duverger O, Lee D, Hassan MQ, Chen SX, Jaisser F, Lian JB, et al. Molecular consequences of a frameshifted DLX3 mutant leading to Tricho-Dento-Osseous syndrome. J Biol Chem. 2008;283: 20198–20208. 10.1074/jbc.M709562200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Choi SJ, Song IS, Ryu OH, Choi SW, Hart PS, Wu WW, et al. A 4 bp deletion mutation in DLX3 enhances osteoblastic differentiation and bone formation in vitro. Bone. 2008;42: 162–171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hassan MQ, Javed A, Morasso MI, Karlin J, Montecino M, van Wijnen AJ, et al. Dlx3 transcriptional regulation of osteoblast differentiation: temporal recruitment of Msx2, Dlx3, and Dlx5 homeodomain proteins to chromatin of the osteocalcin gene. Mol Cell Biol. 2004;24: 9248–9261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Choi SJ, Song IS, Feng JQ, Gao T, Haruyama N, Gautam P, et al. Mutant DLX 3 disrupts odontoblast polarization and dentin formation. Dev Biol. 2010;344: 682–692. 10.1016/j.ydbio.2010.05.499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Duverger O, Zah A, Isaac J, Sun HW, Bartels AK, Lian JB, et al. Neural crest deletion of Dlx3 leads to major dentin defects through down-regulation of Dspp. J Biol Chem. 2012;287: 12230–12240. 10.1074/jbc.M111.326900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Li X, Yang G, Fan M. Effects of homeobox gene distal-less 3 on proliferation and odontoblastic differentiation of human dental pulp cells. J Endod. 2012;38: 1504–1510. 10.1016/j.joen.2012.07.009 [DOI] [PubMed] [Google Scholar]
- 18. Viale-Bouroncle S, Felthaus O, Schmalz G, Brockhoff G, Reichert TE, Morsczeck C, et al. The transcription factor DLX3 regulates the osteogenic differentiation of human dental follicle precursor cells. Stem Cells Dev. 2012;21: 1936–1947. 10.1089/scd.2011.0422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghoul-Mazgar S, Hotton D, Lezot F, Blin-Wakkach C, Asselin A, Sautier JM, et al. Expression pattern of Dlx3 during cell differentiation in mineralized tissues. Bone. 2005;37: 799–809. [DOI] [PubMed] [Google Scholar]
- 20. Chen LS, Couwenhoven RI, Hsu D, Luo W, Snead ML. Maintenance of amelogenin gene expression by transformed epithelial cells of mouse enamel organ. Arch Oral Biol. 1992;37: 771–778. [DOI] [PubMed] [Google Scholar]
- 21. Lv P, Jia HT, Gao XJ. Immunohistochemical localization of transcription factor Sp3 during dental enamel development in rat tooth germ. Eur J Oral Sci. 2006;114: 93–95. [DOI] [PubMed] [Google Scholar]
- 22. Tian H, Lv P, Ma K, Zhou C, Gao X. Beta-catenin/LEF1 activated enamelin expression in ameloblast-like cells. Biochem Biophys Res Commun. 2010;398: 519–524. 10.1016/j.bbrc.2010.06.111 [DOI] [PubMed] [Google Scholar]
- 23. Bartlett JD. Dental enamel development: proteinases and their enamel matrix substrates. ISRN Dent. 2013; 2013: 684607 10.1155/2013/684607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gibson CW. The Amelogenin Proteins and Enamel Development in Humans and Mice. J Oral Biosci. 2011;53: 248–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Wright JT, Hart PS, Aldred MJ, Seow K, Crawford PJ, Hong SP, et al. Relationship of phenotype and genotype in X-linked amelogenesis imperfecta. Connect Tissue Res 2003; 44 Suppl 1: 72–78. [PubMed] [Google Scholar]
- 26. Lezot F, Thomas B, Greene SR, Hotton D, Yuan ZA, Castaneda B, et al. Physiological implications of DLX homeoproteins in enamel formation. J Cell Physiol. 2008;216: 688–697. 10.1002/jcp.21448 [DOI] [PubMed] [Google Scholar]
- 27. Hu JC, Hu Y, Lu Y, Smith CE, Lertlam R, Wright JT, et al. Enamelin is critical for ameloblast integrity and enamel ultrastructure formation. PLoS One. 2014;9: e89303 10.1371/journal.pone.0089303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hu JC, Yamakoshi Y. Enamelin and autosomal-dominant amelogenesis imperfecta. Crit Rev Oral Biol Med. 2003;14: 387–398. [DOI] [PubMed] [Google Scholar]
- 29. Hu Y, Papagerakis P, Ye L, Feng JQ, Simmer JP, Hu JC. Distal cis-regulatory elements are required for tissue-specific expression of enamelin (Enam). Eur J Oral Sci. 2008;116: 113–123. 10.1111/j.1600-0722.2007.00519.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lee HK, Park SJ, Oh HJ, Kim JW, Bae HS, Park JC. Expression pattern, subcellular localization, and functional implications of ODAM in ameloblasts, odontoblasts, osteoblasts, and various cancer cells. Gene Expr Patterns. 2012;12: 102–108. 10.1016/j.gep.2012.02.002 [DOI] [PubMed] [Google Scholar]
- 31. Hassan MQ, Tare RS, Lee SH, Mandeville M, Morasso MI, Javed A, et al. BMP2 commitment to the osteogenic lineage involves activation of Runx2 by DLX3 and a homeodomain transcriptional network. J Biol Chem. 2006;281: 40515–40526. [DOI] [PubMed] [Google Scholar]
- 32. Zhang H, Hu G, Wang H, Sciavolino P, Iler N, Shen MM, et al. Heterodimerization of Msx and Dlx homeoproteins results in functional antagonism. Mol Cell Biol. 1997;17: 2920–2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hu Y, Hu JC, Smith CE, Bartlett JD, Simmer JP. Kallikrein-related peptidase 4, matrix metalloproteinase 20, and the maturation of murine and porcine enamel. Eur J Oral Sci. 2011;119 Suppl 1: 217–225. 10.1111/j.1600-0722.2011.00859.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wright JT, Daly B, Simmons D, Hong S, Hart SP, Hart TC, et al. Human enamel phenotype associated with amelogenesis imperfecta and a kallikrein-4 (g.2142G>A) proteinase mutation. Eur J Oral Sci. 2006;114 Suppl 1: 13–17; discussion 39–41, 379. [DOI] [PubMed] [Google Scholar]
- 35. Nait Lechguer A, Kuchler-Bopp S, Lesot H. Crown formation during tooth development and tissue engineering. J Exp Zool B Mol Dev Evol. 2009;312B: 399–407. 10.1002/jez.b.21256 [DOI] [PubMed] [Google Scholar]
- 36. Hu JC, Simmer JP. Developmental biology and genetics of dental malformations. Orthod Craniofac Res. 2007;10: 45–52. [DOI] [PubMed] [Google Scholar]
- 37. Morasso MI, Grinberg A, Robinson G, Sargent TD, Mahon KA. Placental failure in mice lacking the homeobox gene Dlx3. Proc Natl Acad Sci U S A. 1999;96: 162–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(DOC)
The data represent mean ± SD of three independent experiments, each performed in triplicate.
(TIF)
The data represent mean ± SD of three independent experiments, each performed in triplicate.
(PNG)
As, antisense; S, sense.
(PNG)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.