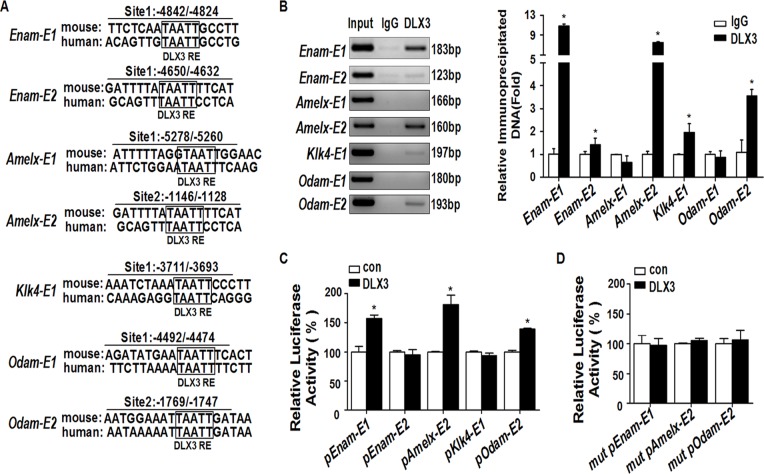
Fig 4. DLX3 directly transactivates the expression of Enam, Amelx, and Odam by binding to the enhancer regions.
(A) Bioinformatic analysis was performed on 6000 bp 5’-flanking regions of Enam, Amelx, Klk4, and Odam. 1 or 2 conserved DLX3 response elements were separately found on the four genes (translation start site defined as +1 bp). RE, response element. (B) ChIP assays determined whether DLX3 was recruited to the predicted enhancer sites. Left panel: gel images of PCR products, representative of three independent experiments. Right panel: statistics of the PCR results. Values are from three independent experiments, and shown as mean ± SD. *P <0.05 vs. IgG group. (C) Luciferase assays were performed to evaluate the impact of DLX3 on the transcriptional activity of each constructed luciferase reporter, which contained enhancer sequences of specific potential DLX3 response elements. Data are compared with the control group (con), and presented as mean% ± SD. *P <0.05. pEnam-E1 represents pGLEnam-E1, etc. (D) Mutations of DLX3 binding sites reduced the activation effect of DLX3 on the transcriptional activity of the pGLEnam-E1, pGLAmelx-E2, and pGLOdam-E2 reporters. The data represent the mean% ± SD of three independent experiments, each experiment performed in triplicate. *P <0.05 vs. the control (con) group. mut pEnam-E1 represents Mut pGLEnam-E1, etc.