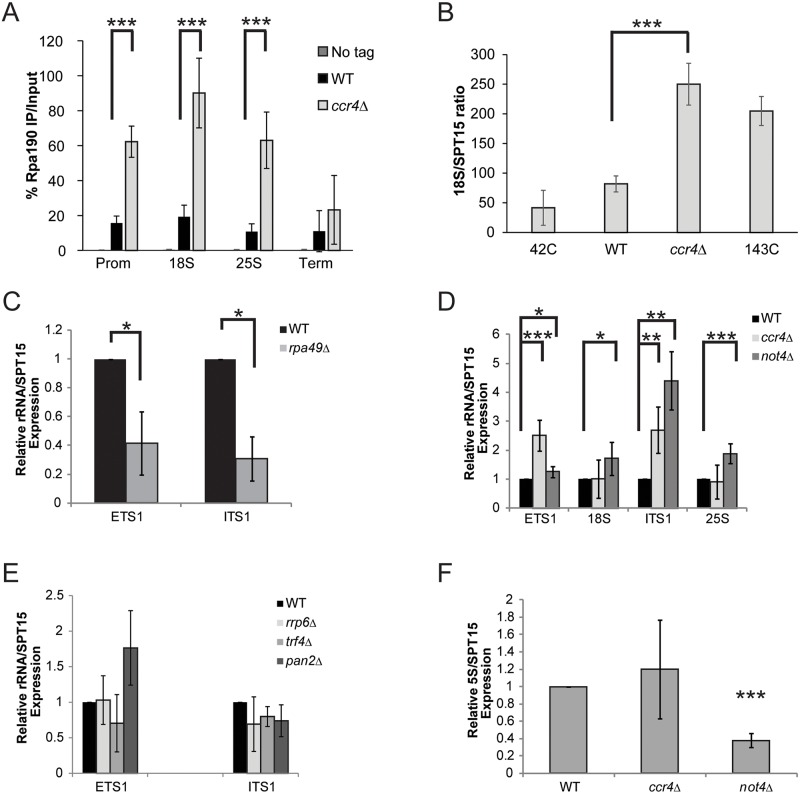
Fig 2. Ccr4-Not disruption increases Pol I transcription.
(A) ChIP for the Pol I catalytic subunit (Rpa190-6XHA) in the no tag, wild-type (WT) and ccr4Δ backgrounds. The average and SD of five independent experiments are shown. (B) 18S/SPT15 ratio from strains 42C (NOY886), 143C (NOY1051), wild-type and ccr4Δ. The average and SD of three independent experiments are presented. (C) ETS1 and ITS1 rRNA expression in wild-type and rpa49Δ. Data are the average and SD of three independent experiments. (D) Expression of the indicated rRNA sequences in WT, ccr4 Δ, and not4 Δ. Data represent the average and SD of five independent experiments. (E) ETS1 and ITS1 rRNA expression from wild-type and the indicated RNA processing mutants. The average and SD of four experiments are shown. (F) Level of 5S rRNA in WT, ccr4 Δ, and not4 Δ. Data are the average and SD of five independent experiments. Student’s t-test determined statistical significance. *- p< 0.05; **-p< 0.01; ***- p< 0.005.