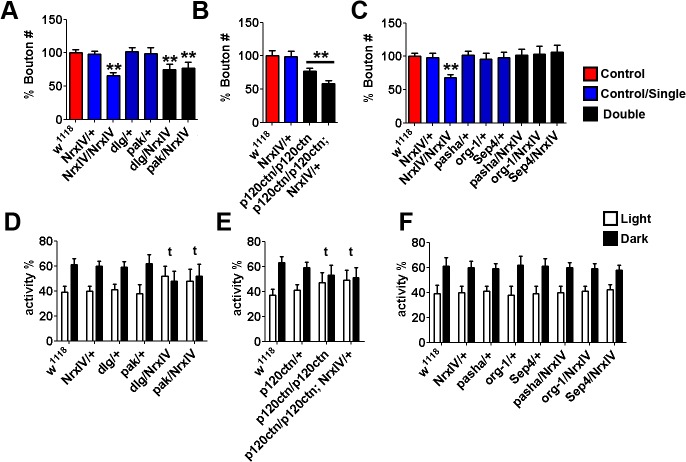
Fig 6. Selective genetic interactions observed between the Drosophila orthologues of ASD candidate genes and Neurexin IV.
A sensitised background of Neurexin IV (NrxIV/+), the orthologue of the autism gene CTNAP2, was used to look for interactions between NrxIV and the ASD candidate gene orthologues dlg, pak, p120ctn, pasha, and org-1. A. dlg/+, pak/+ and NrxIV/+ heterozygous mutants have no significant change in NMJ morphology over w 1118 controls. However, dlg/NrxIV and pak/NrxIV crosses both displayed reduced bouton numbers (n>20 Kruskal-Wallis test, ** P<0.01). B. Significant NMJ morphology changes are also seen for the NrxIV cross with the homozygous p120ctn mutant cross, when compared to the homozygous p120ctn mutant cross alone (n>20 Kruskal-Wallis test, ** P<0.01). C. No NMJ morphology changes were observed when pasha and org-1, both from human de novo loss 12239_chr22_loss_17249508_l, were crossed to NrxIV/+. D. to F. Circadian rhythm analyses of models in Panels A, B and C support observed genetic interactions: dlg/NrxIV, pak/NrxIV transheterozygotes and the NrxIV cross with the homozygous p120ctn mutant flies lost the dark bias, and displayed no significant difference between light/dark sleeping patterns (t), while pasha/NrxIV and org-1/NrxIV transheterozygotes displayed no abnormal circadian phenotype.