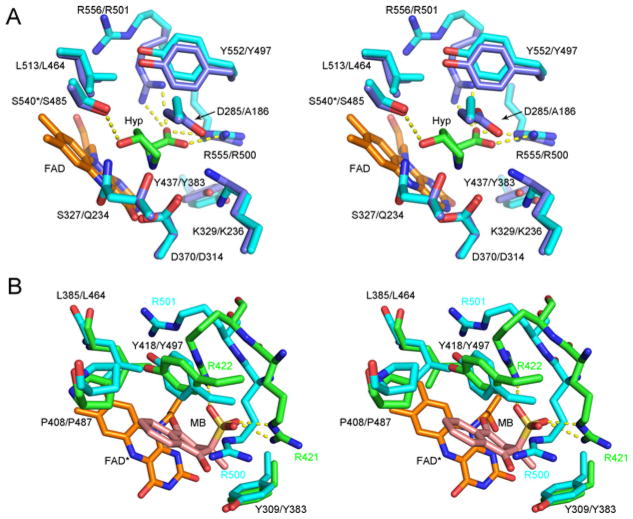
Figure 7. Comparisons of the human PRODH2 model active site to different ligand complexes of enzymes with PRODH domains.
(A) Hyp complex of the Y540S variant of E. coli PutA. The first residue number is that of E. coli PutA, while the second is for hPRODH2. The carbons atoms of E. coli PutA and hPRODH2 are coloured light blue and cyan, respectively. The * indicates the location of the Y540S mutation in Ec PutA. Portions of the FAD molecule have been omitted for clarity. PDB 3E2Q. (B) Menadione complex of GsPutA. Treatment of GsPutA with N-propargylglycine resulted in an imine linkage between the N5 atom of the reduced flavin (FAD*) and the Lys203 (not shown for clarity). The carbon atoms of GsPutA and hPRODH2 are coloured green and cyan, respectively. The first residue number indicated is that of GsPutA; for those cases where the positions of the side chains diverge, the residue label colour corresponds to the model in which they are contained. PDB 4NMF.