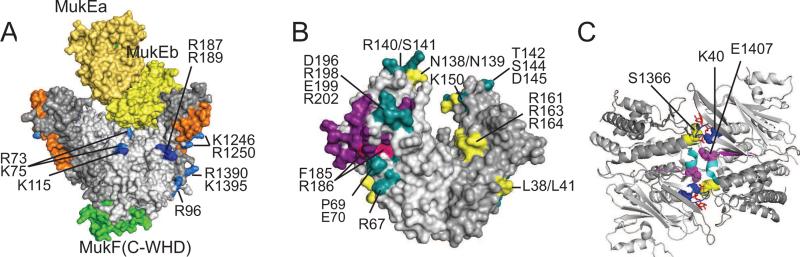
Figure 3.
Architecture of crystallized MukBEF fragments. (A) Crystal structure of dimerized MukB head in complex with a fragment of MukF and a dimer of MukE (PDB 3EUK). The N- and C-terminal domains of MukB are shown in light and darker grey, respectively, MukF is green and the two MukEs are in two shades of yellow. The stems of the long coiled coils are shown in orange. Amino acids involved in DNA binding are blue (essential) or light blue (reduced affinity). Amino acids are numbered according to their position in the E. coli MukB. Note that no MuB-MukE contacts hold MukE above MukB head, where it blocks the DNA binding site.
(B) Crystal structure of MukE (PDB 3EUH) with known mutations. The two MukE monomers are shown in two shades of gray. Purple marks the interface with MukF. Also shown are non-essential amino acids (yellow), essential amino acids found on the MukE-MukF interface (red) and essential amino acids elsewhere (green).
(C) Organization of the ATPase site (PDB 3EUK). The dimeric MukB head viewed from the top with two bound ATPγS (red). Shown are the conserved ATPases regions, including Walker A (blue), C-motif (signature; yellow), Walker B (magenta) and D-loop (cyan). The N- and C-terminal domains are shown in light and darker shades of grey.