Figure 6. Chirality of MukB-DNA interactions.
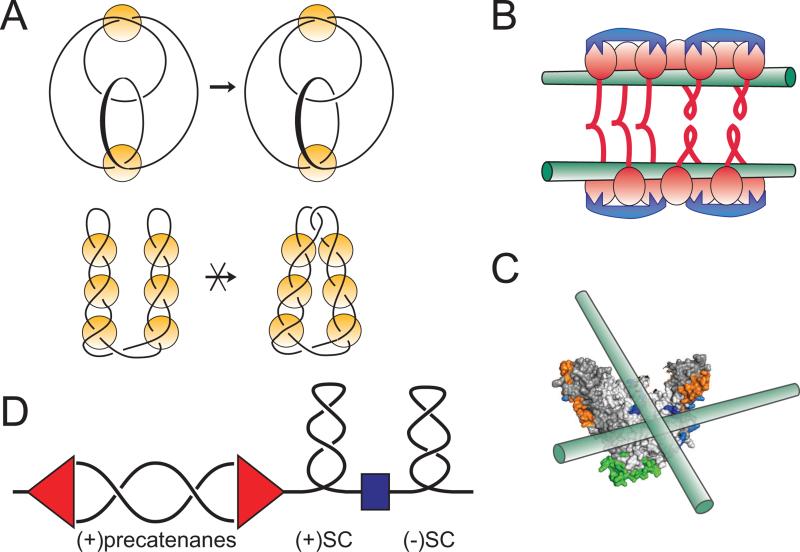
(A) Knotting patterns expected on looped and interwound DNA substrates in topo-2 coupled assay. Knots are often classified according to the minimal number of crossings in a projection of the knot onto a flat surface [Rolfsen, 1976]. The simplest knots, trefoils, contain three irreducible crossings and can exist as either right- or left-handed enantiomers. Condensins promote formation of (+) trefoils, which reveals right-handed DNA looping (top diagram). In contrast, topo-2 reaction on an interwound superhelix would produce twist family knots with multiple irreducible crossings (bottom diagram). Yellow circles depict proteins that capture DNA crossings that were either spontaneously formed in the interwound DNA superhelix (bottom) or comprise the base of protein-induced DNA loops (top).
(B and C) The high preference of MukB for right handed crossings could be explained by chiral binding of DNA to different MukBs from the same cluster (B) or the same MukB head (C).
(C) Pre-catenane formation around replication fork. Progression of the replication fork (red) induces waves of positive supercoiling, whose propagation is limited by domain boundaries (blue). Negative supercoiling ahead of the fork is restored by DNA gyrase. Failure to achieve that results in positive supercoiling which migrates into the replicated region in the form of pre-catenanes. Recruitment of topo IV to this region might facilitate their removal and further progression of DNA replication.