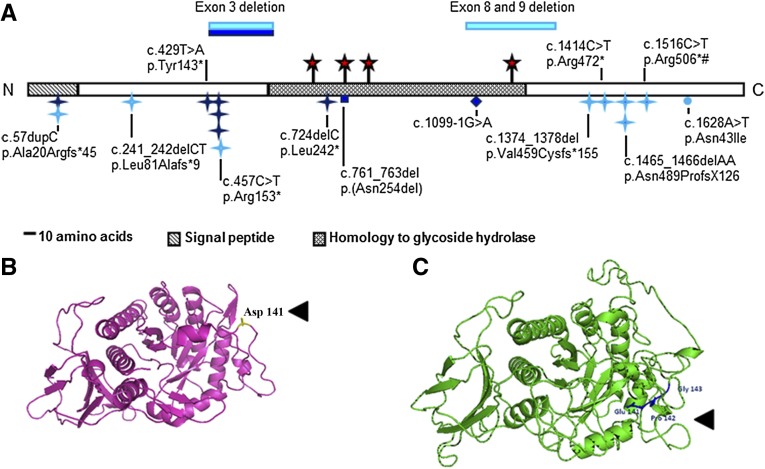
Figure 1.
HPSE2 mutations in UFS and the variant in primary VUR. (A) Schematic of heparanase 2 showing location of mutations. Dark blue indicates mutations described in this report. Light blue indicates mutations from previous reports. Blue stars, nonsense or frameshift mutations; circle, missense mutation; diamond, splice-site mutation; red stars, predicted N-glycosylation sites; #, founder mutation in Ochoa’s Columbian cohort. Domains were predicted by Pfam and SignalP. N and C, the proteins amino and carboxy terminals, respectively. (B and C) Wild-type full length wild-type heparanase 2 protein (B) and the c.422_423insGCCCGG-p.Asp141delinsGluProGly variant (C). The heparanase 2 sequence was aligned to 51 α-L-arabinofuranosidase with ClustalW, and the structural model was generated using PHYRE2 (http://www.sbg.bio.ic.ac.uk/phyre2/html/page.cgi?id=index).