Figure 2.
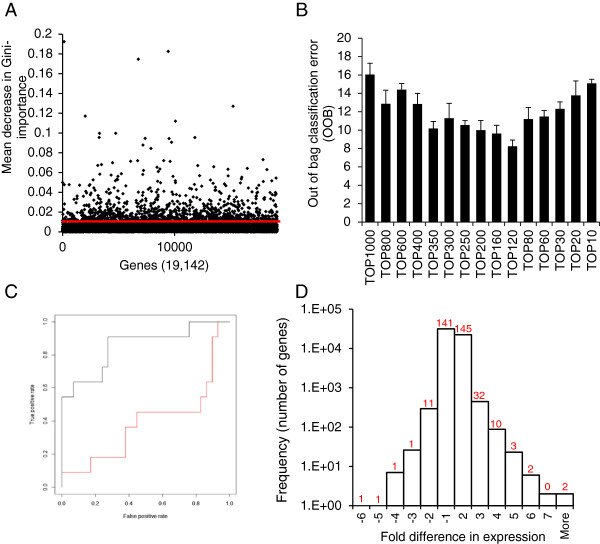
Identification of the core co-acting gene set. (A) Gene ranking by Gini-importance. A singular “best” probeset for each gene was used to grow 10,000 classification trees. The importance of each gene in classifying cell lines as sensitive or resistant to TRAIL was measured by mean decrease in Gini-importance in the training dataset. The probesets above the red line represent the top 5th percentile retained for further analysis. Only genes with Gini-importance value higher than zero were plotted. (B) The top 350 genes predict TRAIL-responsiveness with high accuracy. From the top-ranked 1000 genes, the lowest ranked genes were stepwise removed (by units of 100 and then 10) and the performance of the remaining gene-set was determined by calculating the out of bag classification error (OOB) (stepwise 10-gene unit removal between top 300-top 200 genes had no effect and thus it is not shown on the graph). (C) Validation of the prediction accuracy of the 350 co-acting genes. The area under the receiver operator curve (AUC) was calculated as a measure of the models specificity and sensitivity in the independent test dataset on the dataset (black line, AUC = 0 · 85) as well as after swapping the sensitivity values of a randomly-selected 50% of the cells lines (red line, AUC = 0 · 48). The graph shows the AUC. This is a representative graph from 100 repeats of random permutations. (D) The 350 co-acting genes are not identified by differential expression analysis. A histogram displaying the gene distribution based on fold difference in expression between TRAIL sensitive and resistant cell lines. The number of genes from the panel of 350 co-acting genes falling in the individual fold difference ranges on the histogram is indicated by the numbers above each column.
