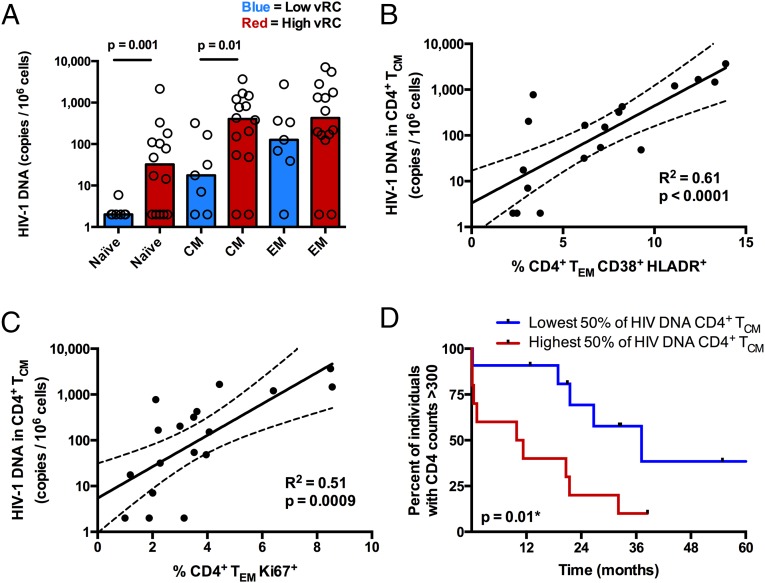
Fig. 6.
Viral RC correlates with the burden of HIV-1 viral DNA in CD4+ TCM and TN. (A) Cryopreserved PBMCs isolated 3 mo postinfection (n = 21) were stained for markers to delineate memory phenotypes, and CD4+ T cells were sorted into three distinct populations, TN (CD27+, CD45RO−), TEM (CD27+/−, CD45RO+, CCR7−), and TCM (CD27+/−, CD45RO+, CCR7+). Cell-associated HIV-1 DNA was quantified by real-time PCR amplification of the HIV-1 subtype C integrase gene. Cell copy number was normalized based on real-time PCR amplification of the human albumin gene. (A) Bar graphs depicting the magnitude of viral burden in TN, TCM, and TEM between individuals infected with either low-RC or high-RC viruses. (B and C) In a majority of the same individuals (n = 18), a fraction of unsorted PBMCs were stained for markers of cellular activation and proliferation. (B) Correlation between the magnitude of viral burden in CD4+ TCM and the percentage of activated (CD38+/HLA-DR+) CD4+ TEM. (C) Correlation between the magnitude of viral burden in CD4+ TCM and the percentage of proliferating (Ki67+) CD4+ TEM. Correlation statistics were generated using linear regression. Solid lines indicate trend lines, and dashed lines represent 95% confidence bands. (D) A Kaplan–Meier survival analysis (with an endpoint defined as CD4+ T-cell counts <300) depicts the effect of viral burden in CD4+ TCM on total CD4+ T-cell decline. Kaplan–Meier survival statistics were generated from the log-rank test.