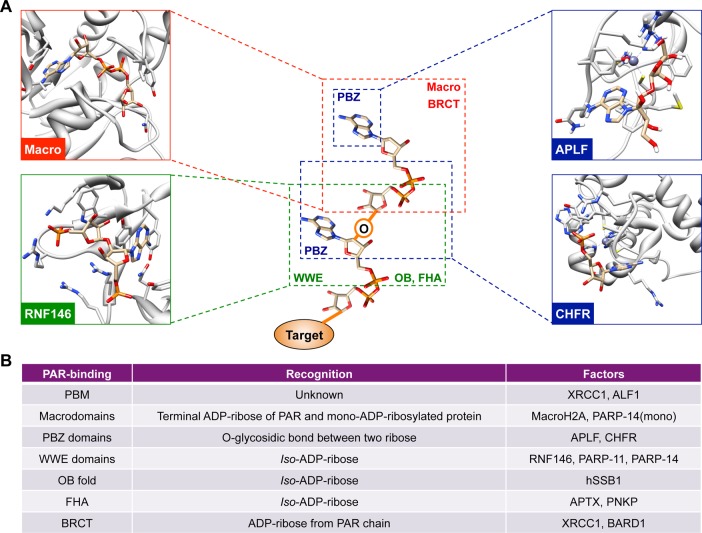
Figure 8.
Recognition of PAR chains by PAR-binding modules. (A) PAR-binding proteins utilize various PAR-binding modules to recognize PAR. The PBZ domain (blue) uses a zinc-coordinated fold that recognizes the α(1 → 2) O-glycosidic bond between two ribose units.10 Solution structure of the first PBZ domain in a complex with ribofuranosyladenosine (upper right panel, PDB: 2KQD)217 and CHFR bound to P(1)P(2)-diadenosine 5′-pyrophosphate (lower right panel, PDB: 2XOY)218 are shown as examples. The macrodomain binds to the terminal ADP-ribose residue of PAR (red, upper left panel, PDB: 2BFQ)78a or mono-ADP ribosylated protein,219 and the WWE domains recognize the iso-ADP-ribose residue (green). Human RNF146 WWE domain in complex with iso-ADP-ribose is shown as an example (lower left, PDB: 3V3L).52 (B) A table summarizing different PAR-binding modules. PAR-binding motifs (PBM) are short amino acid sequences found in PAR-binding proteins such as XRCC1.10 The OB fold is a ssDNA- or RNA-binding motif in prokaryotes and eukaryotes; however, the OB fold of human SSB1 recognizes iso-ADP-ribose.50 BRCT and FHA domain also interact with PAR by recognizing ADP-ribose or iso-ADP-ribose unit from PAR chain, respectively.49