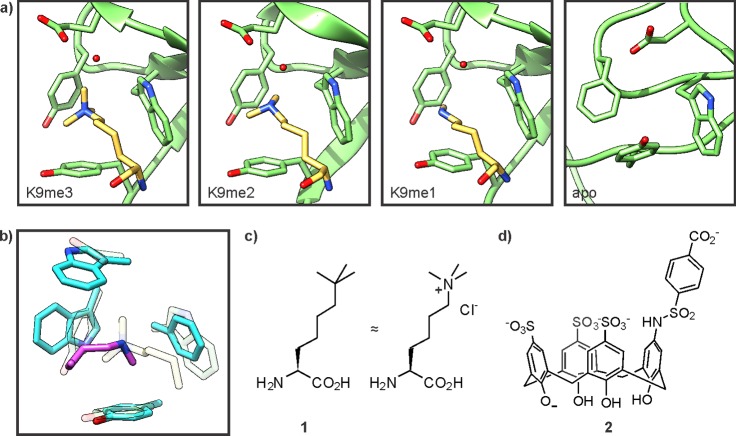
Figure 8.
Recognition of methyllysine. (a) Structures of the HP1 chromodomain in complex with methyllysine residues (pdb codes for K9me3, 1kne; K9me2, 1kna; K9me1, 1q3l) or in its apo form (right, pdb code: 1ap0). The CD is depicted in green, the ligand in yellow. Note that the apo-structure was solved with murine HP1 while the liganded structures were obtained from drosophila HP1, which contains a Tyr residue in place of Phe45. (b) Selective recognition of lower methylation states by the chromodomain of MSL3 (pdb code: 3m9p). The CD is depicted in cyan, the ligand in pink. For comparison, the corresponding residues in the HP1 CD are indicated in pale rendering. (c) Structure of tert-butylnorleucine (1), a trimethyllysine isostere. (d) Structure of a calix[4]arene receptor (2) for methyllysine-containing peptides.