Figure 4.
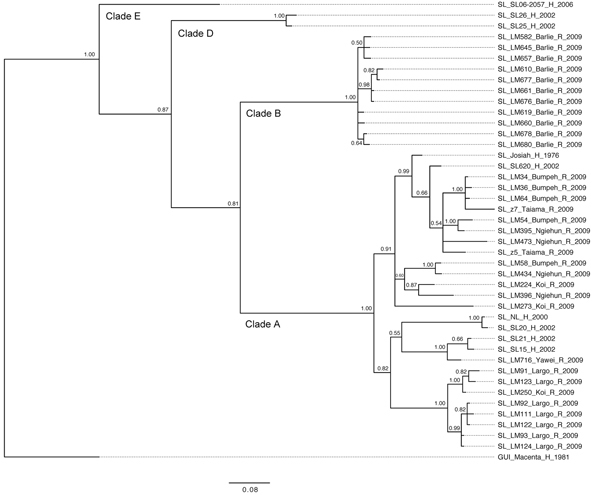
Phylogenetic analysis of Lassa virus (LASV) isolates from Sierra Leone based on partial polymerase (L) gene sequences. The homologous L fragments of 373 nt were aligned. The isolate Z-158, which originated from Macenta district in Guinea, was used as outgroup based on the previous phylogenetic analyses to root the tree. The 50% majority rule consensus tree was estimated by using Bayesian Inference method implemented in MrBayes software (32) using the Tamura 3-parameter substitution model with and a fraction of evolutionary invariant sites. The strain labels contain information on the country of origin (SL, Sierra Leone; GUI, Guinea), strain designation, village or town of origin, type of isolate (H, human; R, rodent), and year of isolation. The numbers next to the branches indicate the posterior probability of particular clades. The clades as defined in this study (clades A, B, D, and E) are also indicated next to the appropriate branches. Scale bar indicates substitutions per site.
