Fig. 2. Genetic evidence that a canonical Sty1 pathway, Int6/eIF3e and Gcn2 specifically promote oxidative stress response during histidine starvation.
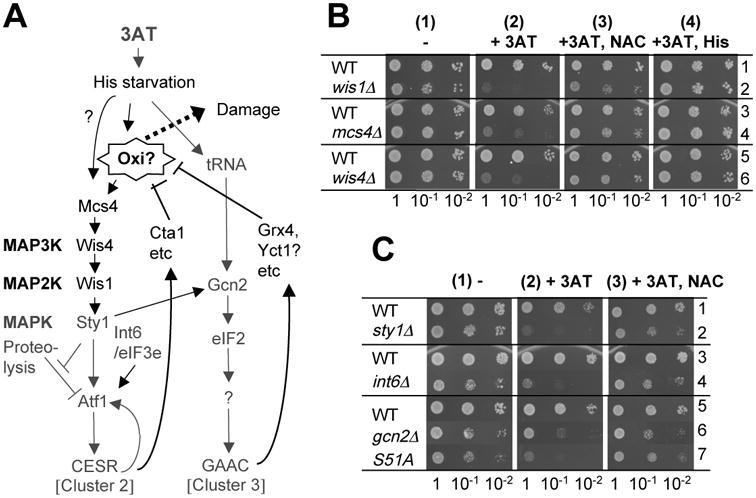
(A) Model of 3AT-induced Sty1 and Gcn2 pathways in fission yeast. 3AT-induced histidine (His) starvation activates not only the Gcn2-dependent pathway (cluster 3, right column) but also the Sty1-dependent pathway (cluster 2, left column). Here it is proposed that at least one of the Sty1 activating signals is endogenous oxidation (Oxi), which would cause cellular damage (Damage) if the products of both the pathways cannot antagonize the oxidative reagents (stopped bars towards Oxi?). This model was modified from the original model described in Ref. 12. Dark gray letters and lines indicate a part of the pathways suggested by previous studies 12. Black letters and lines indicate a part of the pathway suggested from this study. (B-C) Effect of NAC on the 3AT-sensitvity of different mutants. Cultures of the indicated mutants and their wild-type (WT) controls were diluted to A600=0.15, and 5 ml of this and 10-fold serial dilutions were spotted onto EMM plus adenine agar plates supplemented without (panels 1) or with 5 mM 3AT (panels 2), 5 mM 3AT and 20 mM NAC (panels 3), or 5 mM 3AT and 10 mM histidine (His) (panel 4 in A). However, for rows 5 and 6 in Fig. 2B, we used 8 mM 3AT, 40 mM NAC, and 16 mM histidine. Plates were incubated for 4 (panels 1 and 4) and 6-7 (panels 2 and 3) days. Here we used minimal medium without amino acid supplements to promote incorporation of NAC and histidine. Strains used in (B) are JP3 (row 1), JP260 (row 2), KAY793 (row 3), KAY794 (row 4), KAY456 (row 5), and KAY852 (row 6), whereas in (C), we used KAY641 (rows 1 and 3), KAY640 (row 2), KAY647 (row 4), WY764 (row 5), KAY406 (row 6), and JP436 (row 7) (Table 1).
