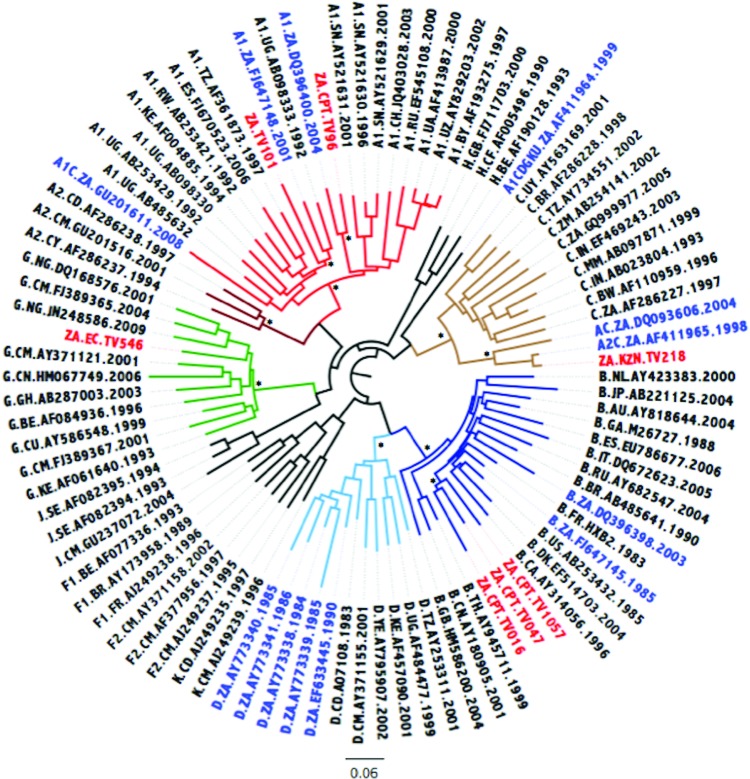
FIG. 1.
Maximum likelihood (ML) tree inference of the near full-length genome (NFLG) concatenated sequence alignment. The ML tree inferred in phylogenetic analysis (phyML) contains the seven newly sequenced Tygerberg Virology (TV) isolates, HIV-1 reference strains, and 13 previously characterized non-subtype C strains from South Africa. The evolutionary distances were computed using the GTR model of nucleic acid substitution with an estimated Gamma shape parameter. The genetic distance is displaying in the scale bar at the bottom of the figure, while the major different clades of HIV-1 Group M have been highlighted. The sequence IDs of South African strains have been marked with the 7 newly genotyped isolates marked in red and the 13 previously genotyped isolates marked in blue. Bootstrap support values for the internal branches for each major clade are shown with an asterisk and indicate support higher than 90%. Color images available online at www.liebertpub.com/aid