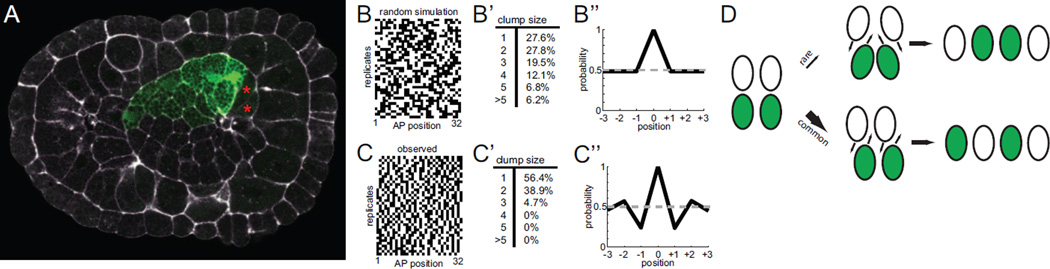
Figure 2. Analysis of primary 16-cell clones.
A) 16-cell primary clone at the onset of mediolateral intercalation showing that the entire right side of the primary notochord primordium is labeled. Not all 16 cells are visible in this focal plane. The right secondary notochord cells have not yet undergone their final division and are marked with red asterisks. Bra>GFP is shown in green and phallacidin staining in white. B) Simulated 16-cell mosaic expression patterns under the hypothesis that notochord intercalation is completely random. C) Mosaic expression patterns for 39 different embryos that showed expression in 16/32 primary notochord cells. The 39 replicates are shown from top to bottom. The AP axis of the notochord is shown from left to right. B’,C’) Quantification of the clumping of labeled cells into groups of different sizes. In the experimental data, most of the labeled cells are singletons (a single labeled cell flanked by two non-labeled cells) or in clumps of only two labeled cells. B’’,C’’) Correlation analysis. A sliding window approach was used to correlate the GFP positive/negative status of each cell with that of its anterior and posterior neighbors. The X axis indicates the probability that cells at varying distances will have the same GFP expression status as the cell at the middle of the window. D) Cartoon diagram indicating two major classes of cell intercalation event. Two cells from one side of the embryo can intercalate between two cells from the other side of the embryo (top). Alternatively, a single cell from one side of the embryo can intercalate between two cells from the other side of the embryo (bottom). The clump size analysis and correlation analysis suggest that the latter possibility is most common.