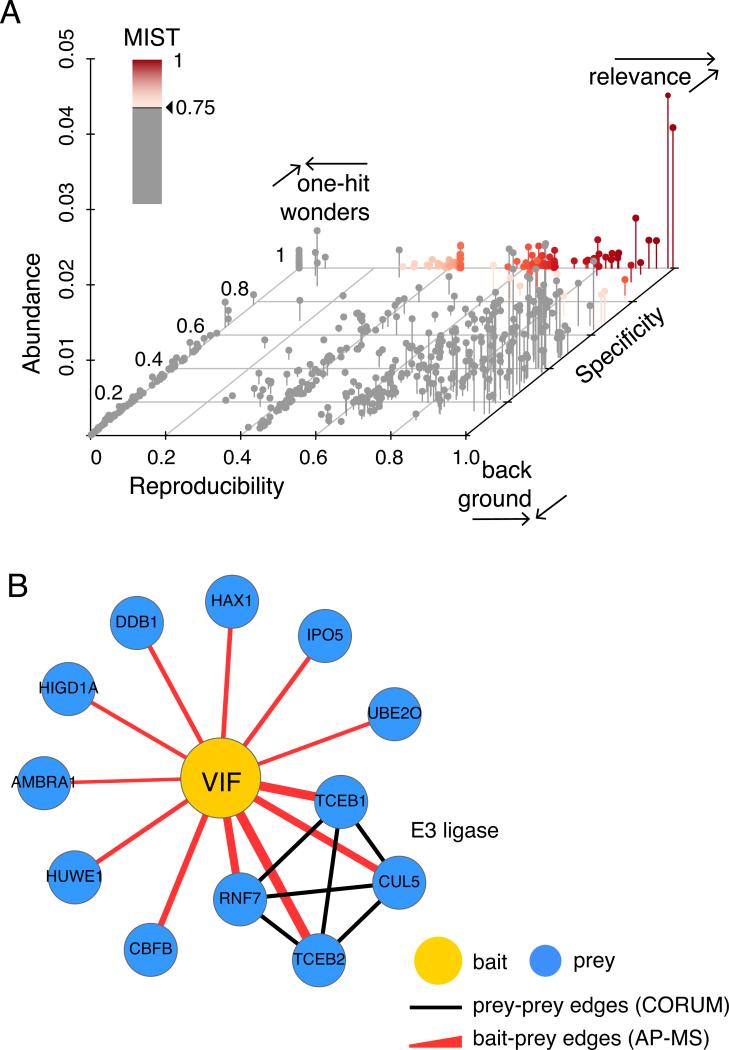
Figure 4.
A) A 3d scatterplot illustrating three different areas in the three-dimensional MiST feature space: (1) biologically relevant interactions (red gradient, MiST scores > 0.75) are specific and reproducible with variable abundance (2) non-specific, reproducible interactions are often background proteins and (3) specific, irreproducible interactions are often one-hit-wonders including contaminants and MS artifacts B) The scored bait-prey list at a high MiST score threshold visualized as a protein interaction network with Cytoscape. Red edges depict interactions identified by AP-MS while black edges are mined from the CORUM database. Edge width corresponds to the MiST score. High MiST scores for multiple subunits of a described complex add confidence to observed AP-MS interactions.