Abstract
The amount of genetic and genomic information obtainable from the human fetus during pregnancy is accelerating at an unprecedented rate. Two themes have dominated recent technological advances in prenatal diagnosis: interrogation of the fetal genome in increasingly high resolution and the development of non-invasive methods of fetal testing using cell-free DNA in maternal plasma. These two areas of advancement have now converged with several recent reports of non-invasive assessment of the entire fetal genome from maternal blood. However, technological progress is outpacing the ability of the healthcare providers and patients to incorporate these new tests into existing clinical care, and further complicates many of the economic and ethical dilemmas in prenatal diagnosis. This review summarizes recent work in this field and discusses the integration of these new technologies into the clinic and society.
A new era of prenatal genomic diagnosis: deeper, faster, and risk free
The concept of the fetus as a genetic entity distinguishable from the pregnant woman is a relatively recent one, dating from 1966 when the first fetal karyotype (see Glossary) was obtained from cultured amniotic fluid cells [1]. At the time, this was recognized as a significant breakthrough in improving the genetic counseling of pregnant women at high risk of having children with chromosome abnormalities. For several decades, the amount of information available to women through prenatal diagnosis has remained almost unchanged, limited to sonographic studies of fetal anatomy and microscopic studies of the fetal metaphase karyotype. Recently, however, great strides have been made in the technologies available for prenatal diagnosis, as exemplified by the ability to sequence the entire fetal genome from maternal blood. Here, we review the recent changes in prenatal screening and diagnosis for genetic disorders and discuss the prospects and challenges created by this progress. A brief overview of the current state of practice is provided in Box 1 and Figure 1.
Box 1. Current state of practice.
Screening for fetal chromosome abnormalities
Aneuploidy is an important cause of perinatal morbidity, mortality, and developmental delay. Trisomy 21 (Down syndrome) is the most common live-born chromosomal abnormality. The risk of a fetus being affected by trisomy 21 is strongly associated with advanced maternal age. It is now standard of care for clinicians to offer Down syndrome screening to pregnant women of all ages [53]. Prenatal screening, previously performed during the second trimester with maternal serum markers, is now predominantly performed during the first trimester, using a combination of ultrasonographic and maternal serum markers. Ultrasonographic fetal nuchal translucency measurement at 11–13 weeks’ gestation [54] and maternal serum screening with free beta-human chorionic gonadotropin (hCG) and pregnancy-associated plasma protein-A detects 85–90% of trisomy 21 with a screen-positive rate of 5% [55,56]. Risk results for trisomy 18 and 13 are also available from first trimester-combined screening protocols. Most women who have positive screening results will undergo invasive testing and will have a euploid fetus. Thus, many invasive tests that are performed are unnecessary.
Diagnosis of fetal chromosome abnormalities
Women at increased risk of carrying a fetus with aneuploidy are typically offered a definitive diagnosis with an invasive procedure. CVS at 11–14 weeks or amniocentesis after 15 weeks’ gestation are the standard methods of obtaining fetal cells for direct assessment of fetal karyotype. Both of these techniques involve a risk of procedure-related miscarriage of approximately one in 300 [57]. For this reason, invasive testing is usually reserved for those women at high risk of fetal aneuploidy, as ascertained by a screening test result, previous history of an affected pregnancy, or advanced maternal age alone.
Classical cytogenetic analysis is highly accurate, but has the limitation of a 7–10-day turn-around time due to the requirement for cell culture [58]. Rapid methods of aneuploidy detection targeted at the most commonly involved chromosomes include fluorescence in situ hybridization, quantitative fluorescent (qf)-PCR, and multiple ligand-dependent probe amplification (MLPA) [59–61]. Due to the advantages of lower cost and automation, qf-PCR has evolved to replace traditional metaphase karyotyping as a stand-alone method of rapid aneuploidy detection in some regions [62]. Molecular karyotyping with chromosomal microarrays is discussed in detail in the main text.
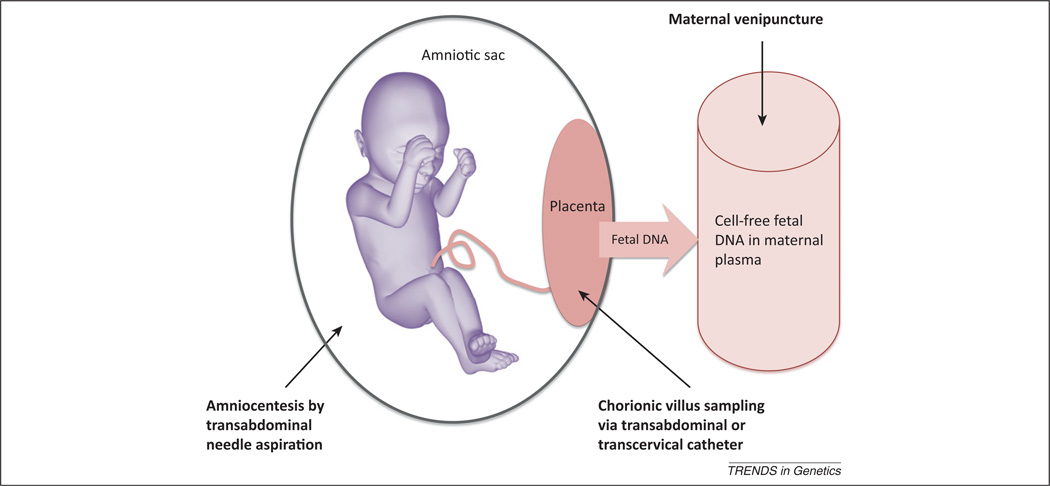
Figure 1.
Sampling methods for genetic assessment of the fetus. Invasive methods carrying a risk of miscarriage include amniocentesis and chorionic villus sampling. Sampling of cell-free fetal DNA in maternal plasma is non-invasive and risk free to the fetus.
Chromosome microarrays
The first prenatal diagnosis of an abnormal karyotype, a balanced translocation, was reported in 1967 [2], which was shortly followed by the first prenatal diagnosis of trisomy 21 (Down syndrome) in 1968 [3]. For both clinical and technical reasons, prenatal diagnosis has historically focused on fetal chromosome abnormalities, which are an important cause of perinatal morbidity and mortality, and are relatively easily detected from cultured fetal cells using standard cytogenetic techniques. Diagnosis during early pregnancy gives women the choice of terminating an affected pregnancy, or continuing to birth with better preparation for the postnatal needs of the child.
Classical cytogenetics (i.e., manual microscopic examination of banded metaphase chromosomes) is now rapidly being superseded as the gold standard of chromosomal assessment. A 2010 international consensus statement recommended that chromosome microarrays (CMAs) replace metaphase karyotyping as the first-tier test for children with unexplained multiple congenital anomalies or developmental delay [4]. CMAs detect genomic gains and losses by hybridizing fluorescently labeled sample DNA onto targets with known genomic coordinates that are fixed to a solid support, such as a glass slide. The relative signal intensity ratio of the sample DNA to a reference sample or database then allows chromosomal gains or losses to be detected. CMAs detect genomic imbalances at a much higher resolution than standard karyotyping (50–100 kb compared with 5–10 Mb [5]) and have a shorter turnaround time because there is no requirement for cell culture. Different types of CMA platform have been used in prenatal diagnosis, including array comparative genomic hybridization using bacterial artificial chromosomes or synthetic oligonucleotide targets, and SNP arrays [6].
A Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD)-sponsored multicenter trial has now provided the first prospective large-scale report on the use of this tool in the assessment of abnormal fetuses. The NICHD trial comparing the accuracy of CMAs to standard cytogenetic analysis for routine and high-risk prenatal diagnosis found that all autosomal and sex chromosomal abnormalities were successfully detected, except triploidy, which is not usually detectable by CMA [7]. Importantly, 5.8% of pregnancies with a fetal structural abnormality and a normal standard karyotype had a clinically significant deletion or duplication detected by CMA, which is consistent with a prior estimate of an additional 5% genomic imbalance detection rate with CMAs when conventional karyotyping is ‘normal’ in a structurally abnormal fetus [8].
The significant yield of additional information with CMAs compared with karyotyping is driving their adoption as the first-tier test in high-risk prenatal cases, such as those with structural abnormalities or severe growth restriction [6]. Recent guidelines from several international and national societies have advocated the selective use of CMAs, rather than the universal application of this technology for all pregnancies undergoing prenatal testing. Identified barriers to systematic use of CMAs in all pregnancies include the complexity of data interpretation, a paucity of appropriate laboratory expertise, and high cost. The European Society of Human Genetics concluded that arrays were of proven value for investigation of fetal abnormalities and encouraged the establishment of local guidelines for the use of genome-wide array analysis in the prenatal setting [9]. They also recommended that pretest counseling, including written information and parental consent, were essential components of such a diagnostic service. In its 2012 position statement, the Italian Society of Human Genetics recommended that CMAs only be used for specific diagnostic purposes in selected pregnancies and ‘never’ as a substitute for conventional karyotyping [10]. This is in line with the recommendations from the American College of Obstetricians and Gynecologists, which has not yet altered its 2009 recommendation that conventional karyotyping remain the principal cytogenetic tool in prenatal diagnosis [11].
One of the greatest concerns with the use of CMAs in prenatal diagnosis is the complexity of data interpretation and subsequent genetic counseling challenges [12]. Approximately 1% of prenatal CMAs return results of ‘unknown significance’ [8]. The difficulty of distinguishing benign versus pathological copy number variations (CNVs) is exacerbated in the prenatal setting, where only limited phenotype information is available. Features of pathogenic CNVs in pediatric patients have been well described, but these must be interpreted with caution in prenatal samples (Box 2) [4]. Parental DNA may be required for complete interpretation of CNVs, which may not always be available or may create ethical dilemmas regarding paternity. Public reference databases, such as the database of Genotypes and Phenotypes (dbGAP) and the Database of Chromosomal Imbalance and Phenotype in Humans using Ensembl Resources (DECIPHER), can be consulted to interpret CNVs, but this requires specific training and expertise. Additional challenges for counseling include incidental findings, such as the detection of clinically relevant imbalances that are unrelated to the indication for testing or misattributed paternity. All these issues are particularly acute when pregnancy termination is being considered. To address these concerns, many specialists advocate using a targeted rather than a whole-genome approach. These targeted platforms cover genomic regions that are associated with full penetrance for intellectual disabilities, birth defects, or other well-characterized developmental disabilities, reducing and simplifying data interpretation at the expense of the depth of genomic information.
Box 2. Features of pathogenic CNVs.
The features of pathogenic CNVs listed below are adapted from [4].
Primary criteria
Identical CNV inherited from an affected parent.
Expanded or altered CNV inherited from a parent.
Similar to a CNV in an affected relative.
Overlaps a genomic imbalance defined by a high-resolution technology in a CNV database for patients with intellectual disability and/or developmental delay, autism spectrum disorder, or multiple congenital anomalies.
Overlaps genomic coordinates for a recognized deletion or duplication syndrome.
Contains known disease-causing genes.
Gene-rich region.
General findings
Deletion.
Amplification of more than one copy gain.
The NICHD has recently funded a long-term study to follow postnatal outcomes of those fetuses assessed by prenatal CMAs. These data are needed to improve knowledge and provide more accurate genetic counseling to prospective parents. In addition, CMA sequence data collected in a central archive could become a valuable resource to advance knowledge of human development and population health. This is the goal of publicly available registries, such as dbGAP (http://www.ncbi.nlm.nih.gov/gap) and the International Standards for Cytogenomics Arrays Consortium (ISCA; http://www.iscaconsortium.org).
Non-invasive prenatal diagnosis
Non-invasive prenatal diagnosis (NIPD) refers to the overall assessment of fetal health without directly accessing the uterus and, therefore, without the associated risk of miscarriage inherent in amniocentesis or chorionic villus sampling (CVS). NIPD encompasses both ultrasound examination and maternal blood sampling for a wide range of fetal conditions, including anemia and genetic disorders, the latter of which we discuss here. Research in this field has focused on two major themes: (i) the detection of specific fetal disorders caused by paternally inherited genes; and (ii) the detection of fetal aneuploidy. In the subsequent sections, if a test is considered to be diagnostic of a fetal condition, the term ‘NIPD’ is used throughout. Because detection of fetal aneuploidy using maternal blood is not yet diagnostic, ‘non-invasive prenatal testing’ (NIPT) is currently the preferred term.
The greatest successes in NIPD and NIPT for fetal genetic conditions have been achieved using cell-free fetal (cff) DNA in maternal blood. Knowledge has advanced extremely rapidly since its existence was first reported in 1997 [13]. The major source of circulating cff DNA and RNA is the trophoblast, which releases nucleic acids into the maternal circulation within microparticles that protect them from degradation by plasma nucleases. This uniquely accessible fetal material has been intensively investigated for the assessment of single gene mutations, fetal chromosome abnormalities, and even the entire fetal genome.
NIPD of fetal sex and monogenic disorders
Non-invasive methods using circulating cff DNA are an established alternative to invasive testing for the diagnosis of fetal sex and Rhesus (Rh) D antigen status. Prenatal diagnosis of fetal sex is clinically relevant for pregnancies in which the fetus is at risk of X-linked disease, in utero virilization from congenital adrenal hyperplasia, or when ambiguous genitalia are identified by sonographic examination. NIPD using real-time quantitative PCR for the male-specific loci sex-determining region Y (SRY) or testis specific protein, Y-linked 1 (alias DYS14) is reliable for fetal sex from 7 weeks’ gestation, permitting a much earlier diagnosis than is possible by either invasive testing or ultrasound examination. NIPD for fetal sex has an overall sensitivity of 95.4% and a specificity of 98.1% [14]. However, with stringent reporting criteria, the accuracy can be as high as 99.5% in clinical practice, translating into a substantial reduction in the number of women requiring invasive procedures [15].
One of the main technical limitations of the test is the difficulty in establishing a positive control to demonstrate successful isolation of cff DNA in female fetuses. As female fetal sex is not detected directly, but inferred from the absence of Y-specific sequences, a negative result is indistinguishable from failed amplification of cff DNA. Two main approaches to detecting sex-independent unique fetal DNA sequences in maternal plasma have evolved. One is to use informative paternally inherited SNPs that are absent in the maternal genome, an approach used by National Health Service laboratories in the UK [15]. Maternal DNA is analyzed for the presence or absence of a bank of eight bi-allelic polymorphisms or markers. Markers found to be absent from the maternal genome are targeted in maternal plasma and, if at least two of four replicates of any marker are positive, fetal DNA is presumed to be present in the maternal plasma.
An alternative epigenetic method utilizes the methylation status of the promoter region of the tumor suppressor gene Ras association (RalGDS/AF-6) domain family member 1 (RASSF1A) to identify DNA sequences of placental origin in maternal blood [16]. The RASSF1A gene is hypermethylated in the placenta, but hypomethylated in maternal blood cells. Methylation-sensitive endonucleases can be used to digest maternal hypomethylated RASSF1A sequences before amplification, allowing the intact placental RASSF1A sequences to be used as positive controls for the presence of fetal DNA. The clinical utility of this approach has recently been demonstrated in several studies [17,18].
NIPD of fetal Rh blood group is another major clinical application that utilizes cff DNA in maternal blood. The primary aim of testing is to allow early identification of fetuses at risk of hemolytic disease in alloimmunized pregnant women. The basis of this assay is the amplification of the fetal Rh blood group, D antigen (RHD) gene in Rh D-negative pregnant women who would not otherwise have an amplification product. This test first became widely available more than 10 years ago in the UK [19]. Building on the success of the non-invasive approach, Denmark has recently introduced screening of Rh-negative women for a Rh-positive fetus to allow targeted prophylaxis with anti-D immunoglobulin [20]. The published audit showed substantial reductions in both costs and unnecessary exposure of pregnant women to blood products.
Similar NIPD tests are now available for other blood group antigens that can cause fetal hemolytic anemia [21]. Future prospects for NIPD include its continued expansion to cover a greater range of disorders, including hemoglobinopathies [22] and fetal and/or neonatal alloimmune thrombocytopenia [23]. The technology also continues to advance beyond qualitative PCR for specific fetal sequences, to include DNA arrays for common SNPs associated with beta-thalassemia [24] and microfluidics digital PCR for relative mutation dosage in hemophilia [25].
NIPT for aneuploidy using massively parallel sequencing of cell-free DNA in maternal blood
Trisomy 21 is the most common genetic cause of intellectual disability. NIPD of fetal trisomy 21 has been a long-standing goal of prenatal diagnostics. Early attempts using fluorescent in situ hybridization on intact fetal cells isolated from the maternal circulation proved labor intensive and ultimately unsuitable for translation into clinical practice [26]. An alternative approach based on the determination of fetal chromosome dosage from total (maternal and fetal) cell-free DNA fragments in maternal blood has evolved, initially using digital PCR [27,28]. Detection of fetal aneuploidy using cell-free DNA is more difficult than the diagnosis of single gene disorders because the fetal sequences from chromosome 21 are indistinguishable from those of maternal origin. Despite this, fetal trisomy 21 can be detected because a woman carrying a fetus with Down syndrome will have a higher proportion of chromosome 21 DNA fragments in the pool of her total plasma cell-free DNA than an equivalent pregnant woman with a euploid fetus. Approximately 10% of total cell-free DNA in maternal blood originates from the fetus during the first and second trimester [29]; therefore, high precision is required to determine whether more than the expected amount of DNA from chromosome 21 is present. This technique requires millions of DNA molecules to be sequenced, counted, mapped to a reference human genome, and analyzed using sophisticated bioinformatics tools. The recent application of massively parallel sequencing (MPS) technology to this task has made NIPT feasible on a scale suitable for large clinical trials and commercial release.
During the past 18 months, multiple clinical validation studies have been published describing results of NIPT for fetal aneuploidy. The first four independent clinical trials of NIPT of trisomy 21 using MPS were all performed in high-risk populations and demonstrated near-perfect detection rates for trisomy 21 (98.6–100%), with superior sensitivities and specificities compared with standard serum biochemical screening tests [30–33]. In particular, the low false positive rates (<1%) have demonstrated the potential of NIPT to dramatically reduce the invasive testing rate compared with the current standard screening tests, which typically have a 5% screen-positive rate.
At least seven more clinical trials have been published since these initial reports. All of them use some version of sequencing to detect fetal chromosomal disorders beyond Down syndrome, including trisomies 18 and 13, as well as sex chromosome disorders [34–40]. From these studies, it appears that MPS approaches are less sensitive for detecting trisomies 18 and 13 compared with trisomy 21. This difference in performance has been attributed to the relatively low guanine and cytosine contents of chromosomes 18 and 13, because biases in sequencing coverage occur in regions of low or high GC content. Statistical normalization of the sequenced read number to the GC content of the reads removes the bias effect and can improve detection rates of trisomies 18 and 13 from 73% and 36% to 91.9% and 100%, respectively [36]. A recently reported alternative method of overcoming GC bias is single-molecule sequencing, which omits the PCR amplification step used in other MPS platforms that can introduce the GC bias [41].
Despite the technical progress of NIPT, the cost and computational analysis of MPS technology remains a barrier to its widespread use. Selective analysis of cell-free DNA from chromosomes 21 and 18 using a highly multiplexed assay, called digital analysis of selected regions (DANSR), can improve mapping efficiency, sequencing cost, and throughput of MPS [38,40]. In the DANSR method, locus-specific oligonucleotides bind to cell-free DNA sequences unique to the chromosomes of interest. The hybridized oligonucleotides are amplified using universal PCR primers, and the PCR reaction products are sequenced. The depth of sequencing required in this chromosome selective method is less than 5% of that required by whole-genome sequencing (1 million raw reads per subject compared with 25 million), which translates to reduced cost and higher throughput [34,39]. The disadvantage of targeted sequencing is that abnormalities in chromosomes other than 13, 18, and 21 will not be detected.
One of the technical insights gained as a result of these sequencing studies is that the proportion of cell-free DNA of fetal origin in the maternal plasma (e.g., the ‘fetal fraction’) is a major influence on test performance. Samples that contain a low fetal fraction (<4%) are more likely to return an inconclusive result. Fetal fraction varies between individuals and decreases with increased maternal body mass index [42]. It is also possibly increased in twin compared with singleton pregnancies [43]. More information on the factors that influence fetal fraction will be important to assist in the future application and interpretation of MPS results.
NIPT for fetal aneuploidy is currently positioned clinically as an ‘advanced screening test’ rather than a diagnostic test. It is indicated for pregnant women who have been previously identified as being at high risk for carrying an aneuploid fetus and who prefer to avoid or delay an invasive diagnostic test. In a Rapid Response Statement released in October 2011, the International Society for Prenatal Diagnosis recommended confirmation of abnormal MPS results by invasive testing and cautioned against its routine use in low-risk populations [44]. Early experiences in the USA and China have demonstrated that pregnant women and their providers are ready to adopt this technology. The prospect of MPS testing for aneuploidy as a primary screening is an ongoing debate. As insurance companies widen eligibility for the test, it is expected that uptake will increase. However, unintended consequences of NIPT, such as the detection of previously undiagnosed maternal genetic disorders, are likely to emerge as this procedure becomes more common [45].
The great promise of NIPT for fetal aneuploidy is to achieve diagnostic accuracy, thereby replacing invasive testing altogether and making prenatal diagnosis safer. The combined data from the published literature provide an overall estimated sensitivity and specificity of 99% for detection of fetal Down syndrome [46]. That this has been achieved only 15 years after fetal DNA was first discovered in maternal blood is truly remarkable. To achieve 100% diagnostic accuracy will require increased sequencing depth, which will have additional implications regarding costs and turnaround time.
Non-invasive sequencing of the entire fetal genome from maternal blood
Current prenatal screening programs focus on a limited number of common aneuploidies, such as trisomy 21. However, when considered in total, rare Mendelian disorders comprise as much as 1% of all congenital diseases. Non-invasive assessment of the entire fetal genome has the potential to detect these rare disorders without posing any risk of miscarriage, taking prenatal diagnosis to the next level.
It has been known for several years that DNA molecules across the entire maternal and fetal genomes are present in maternal plasma in constant relative proportions [47]. In a 2010 proof-of-concept study, the feasibility of non-invasive assessment of the fetal genome was established using a mother–father–child trio in which the parents were carriers of different beta-thalassemia mutations. In this study, the investigators performed SNP genotyping from maternal and paternal buffy coat samples and chorionic villi [47]. Paired-end sequencing of DNA extracted from maternal plasma to 65-fold genome coverage allowed the investigators to assemble the fetal genome using the paternal genotype and maternal haplotype (deduced from the CVS) as guides. They then scanned the fetal genome to see whether the fetus had inherited the two beta-thalassaemia mutations. The paternal mutation was detected in maternal plasma, confirming fetal inheritance of this mutation. To determine whether the fetus had inherited the maternal mutation, a relative haplotype dosage analysis was performed. A relative excess of the maternal haplotype without the mutation indicated that the fetus inherited the wild type allele and therefore was only a carrier of the paternal mutation.
The haplotype-based approach to determine fetal inheritance has been further developed in two recent studies reporting non-invasive assessment of the fetal genome from maternal plasma. In the first study, the investigators integrated the haplotype-resolved genome sequence of the mother, the shotgun sequence of the father, and the deep sequencing of cell-free DNA in maternal plasma (maternal and fetal) to non-invasively predict the whole-genome sequence of a fetus [48]. Unlike the previous study [47], they did not require an invasively acquired fetal sample (via CVS) to deduce the maternal haplotype. By sequencing complex haploid subsets of maternal genomic DNA while preserving long-range contiguity, they directly determined the phase of 91.4% of 1.9 × 106 heterozygous SNPs into long haplotype blocks. Allelic imbalances in maternal plasma manifesting across the experimentally determined maternal haplotype blocks were used to predict maternal transmission to the fetus. The observation (or lack) of paternal alleles in maternal plasma was used to predict paternal transmission. Candidate de novo mutations in the fetus were identified by analysis of alleles in maternal plasma that were not present in maternal or paternal genomic DNA.
Both of these studies required paternal samples to determine the fetal genome, which is a practical limitation to its clinical application. In a significant recent development, another group reported the non-invasive measurement of the fetal genome using a method that did not require paternal samples [49]. Two pregnant women were assessed, one with a normal fetus and one with a heterozygous deletion on chromosome 22 associated with DiGeorge syndrome. The authors applied the chromosome counting principle used in aneuploidy detection to count haplotypes and individual alleles directly. In contrast to the two previous studies, they determined the maternal haplotype by a method called direct deterministic phasing using three to four single lymphocytes [50]. The maternal heterozygous loci were then used to define the two maternal haplotypes. Shotgun sequencing of the plasma cell-free DNA was performed, and the relative amounts of parental haplotypes were measured by counting the number of alleles specific to each parental haplotype (‘markers’). The paternally inherited haplotypes were reconstructed by detection of paternal-specific alleles, followed by imputation at linked positions using reference haplotypes from the 1000 Genomes project. This method allowed deduction of the inheritance of each parental haplotype and construction of the full inherited fetal genome. Significantly, this haplotyping method was able to identify correctly the deletion on chromosome 22 that the mother transmitted to her fetus. These advances overcome several major technical barriers to the non-invasive assessment of the whole fetal genome, bringing researchers closer to the clinical application of this technology.
Prospects and challenges
Technological progress in prenatal diagnostics has finally overcome the dual challenges of the genetic complexity and the physical inaccessibility of the fetus, making non-invasive prenatal assessment of the entire fetal genome a reality (Figure 2). In the future, it is possible that a woman may have a first-trimester blood test that will inform her as to whether her fetus has a chromosome abnormality and/or dozens of single gene mutations, and/or thousands of polymorphisms. Genetic counseling regarding these results will be a major challenge. Furthermore, the ethical issues that arise in prenatal diagnosis are more acute than in adult or pediatric medicine because the prospective parents are often trying to make decisions about termination of pregnancy based on the results [51]. Although these ethical issues have always been present in prenatal diagnosis, the scale and rapidity of the changes brought about by recent technological advances heightens the concerns, especially given the availability of direct-to-consumer genetic testing in the USA [52].
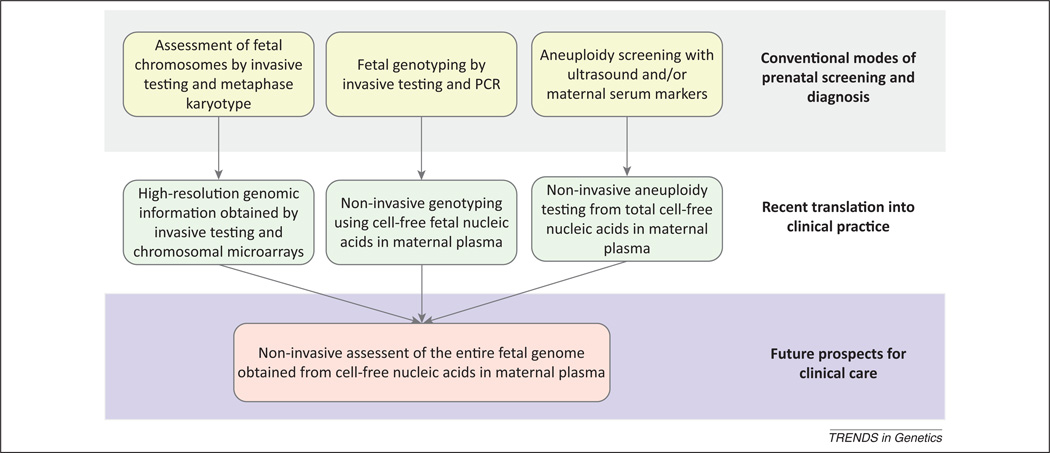
Figure 2.
Progress and prospects for prenatal diagnosis. The future of prenatal diagnosis will see the converging of advances in non-invasive and genome-wide techniques.
Whatever technological platform is eventually used to assess the fetus, whether targeted or whole genome, data interpretation, storage, and management will be major challenges for future clinicians and diagnostic services. It is also clear that these advances are outpacing the ability of clinicians and consumers to absorb their impact on clinical care. The associated challenges in clinical and public education and healthcare economics need to be urgently addressed. Multidisciplinary collaboration between clinicians, patients, academia, and industry will be essential to reap the benefits to scientific knowledge and patient care created by this technology (Box 3).
Box 3. Challenges created by recent advances in prenatal diagnosis.
Challenges for the healthcare system
Provision of adequate counselor and practitioner education about new technologies.
High-quality information provision and genetic counseling for all pregnant women regarding options for antenatal screening and testing.
Standardized guidelines for testing and reporting.
Quality assurance measures for ultrasound assessment, laboratory methods, and risk calculation software.
Improved approaches for dealing with results of uncertain significance and unanticipated findings (such as predisposition for adult-onset diseases, misattributed paternity, and maternal genetic disorders).
Creation of data registries across private and public sectors to better understand the postnatal phenotype of prenatally detected CNVs.
Assessment of the cost-effectiveness of new technologies and appropriate incorporation of these approaches within the existing healthcare system.
Careful partnerships between industry and academia that address financial conflicts of interest, intellectual property disputes, and open data sharing to advance medical knowledge.
Ethical challenges
Better understanding of patient attitudes to new methods of prenatal testing, including the role of cultural background and financial resources.
Overhaul of the informed consent process in the context of unprecedented data generation.
Ensuring equity of access to new standards of care.
Direct-to-consumer testing in NIPD: ensure adequate quality control, provision of appropriate genetic counseling, and psychological support to consumers.
Continued provision of support and resources for women who refuse testing or who continue a pregnancy with a diagnosis of a fetal abnormality.
Finally, more attention needs to be paid to the needs and expectations of women and their partners with regard to prenatal diagnosis. Only then will communities be able to set appropriate priorities for resource allocation and determine the appropriate place of new technologies in prenatal care.
Acknowledgments
L.H. received financial support from the University of Sydney Medical School Albert S. McKern scholarship and the Royal Australian and New Zealand College of Obstetricians and Gynecologists Fotheringham Fellowship. D.W.B.’s time and effort in writing this manuscript was partially supported by the US National Institutes of Health grant HD42053-09.
Disclaimer statement
D.W.B. is the Chair of the Clinical Advisory Board of Verinata Health. In this position, she receives honoraria and stock options. She is also the recipient of a sponsored research grant from Verinata Health.
Glossary
- Amniocentesis
sampling of amniotic fluid via transabdominal ultrasound-guided needle aspiration, typically a 10–20-ml volume taken at 15–20 weeks of gestation. The amniocytes contained in the fluid are cultured and used for determination of fetal karyotype.
- Cell-free fetal (cff) DNA
the free-floating DNA fragments of fetal and/or placental origin present in a biofluid after residual cells have been removed through high-speed centrifugation. The cell-free DNA is contained within microparticles that protect them from degradation by circulating nucleases. Cell-free DNA is also present in body fluids in the non-pregnant state and has been extensively studied in cancer medicine.
- Chorionic villus sampling (CVS)
the biopsy of microgram quantities of placental tissue via transabdominal or transcervical ultrasound-guided needle aspiration, usually performed at 10–13 weeks of gestation.
- Chromosome microarrays (CMAs)
also known as microarray-based genomic copy number analysis or molecular karyotyping. Includes all types of array-based genomic copy number analysis, including array-based CGH (array CGH) and SNP arrays. CMA performs a similar function to G-banded karyotyping but at a higher resolution.
- Copy number variants (CNVs)
structural variations in the DNA of a genome due to variations in the number of copies of one or more sections of DNA. These can take the form of deletions or duplications. CNVs differ from SNPs in that the size of the affected region is larger (up to several megabases). A CNV of ‘unknown significance’ is one that that has not been previously identified in the patient population of a laboratory, has not been described in the medical literature or publicly available databases, and does not contain any known disease-causing genes.
- Haplotype
a set of DNA variations, or polymorphisms, that tend to be inherited together. A haplotype can refer to a combination of alleles or to a set of SNPs found on the same chromosome.
- Karyotyping
the process of determining the number and structure of chromosomes in a eukaryotic cell. In conventional cytogenetic analysis, cultured cells are used to prepare stained metaphase chromosomes for examination under a light microscope (G-banded karyotyping). Chromosomal number, length, position of centromeres, banding pattern, and other physical characteristics are visually assessed by a cytogeneticist.
- Massively parallel sequencing (MPS)
a sequencing method in which many thousands or millions of sequencing reactions are performed in parallel, greatly reducing the cost and increasing the throughput of sequencing.
- Non-invasive prenatal diagnosis (NIPD)
the diagnosis of fetal conditions without invasive sampling of fetal tissue (i.e., via CVS or amniocentesis) and, therefore, without any procedure-related risk of miscarriage. This term is now commonly used for diagnostic tests utilizing cff DNA in maternal plasma. The results of non-invasive diagnostic tests are clinically actionable and do not generally require confirmation by invasive testing (e.g., fetal sex determination).
- Non-invasive prenatal testing (NIPT)
the non-invasive assessment of fetal health, typically using cell-free DNA in maternal blood. This term encompasses non-invasive screening tests that require confirmation with invasive testing, such as fetal aneuploidy assessment. It has become the preferred term for fetal aneuploidy assessment using cell-free DNA in maternal plasma, because this test has not yet reached diagnostic accuracy.
- Nuchal translucency
the fluid-filled space at the back of the neck of the first-trimester fetus that appears echolucent on ultrasound examination. An increased measurement of this space at 11–14 weeks of gestation is strongly associated with an increased risk of trisomy 21 and/or fetal malformations.
- Phasing
the process of determining which alleles (indicated by the variant SNPs) are located together on the same chromosome; conventional methods include using family trio data or aggregating population genotype data.
- Rhesus D antigen
the most immunogenic antigen of the Rh blood group system and the most common cause of hemolytic disease of the fetus and/or newborn. Such disease is preventable by administration of anti-D immunoglobulin to Rh-negative pregnant women to prevent alloimmunization during pregnancy with a Rh-positive fetus.
- Shotgun sequencing
a laboratory technique for determining the DNA sequence of the genome of an organism. Genomic DNA is broken into random fragments that are individually sequenced using the chain termination method to obtain reads. Multiple overlapping reads for the target DNA are obtained by performing several rounds of fragmentation and sequencing. A computer program looks for overlaps in the DNA sequences and uses them to arrange the individual fragments in their correct order to reconstitute the genome.
- SNPs
the most common type of genetic variation. Each SNP represents a DNA sequence variation where a single nucleotide differs between members of a biological species in a given stretch of DNA. SNPs occur approximately once in every 300 nucleotides and can be used as biological markers.
References
- 1.Steele MW, Breg WR., Jr Chromosome analysis of human amniotic-fluid cells. Lancet. 1966;1:383–385. doi: 10.1016/s0140-6736(66)91387-0. [DOI] [PubMed] [Google Scholar]
- 2.Jacobson CB, Barter RH. Intrauterine diagnosis and management of genetic defects. Am. J. Obstet. Gynecol. 1967;99:796–807. doi: 10.1016/0002-9378(67)90395-x. [DOI] [PubMed] [Google Scholar]
- 3.Valenti C, et al. Prenatal diagnosis of Down’s syndrome. Lancet. 1968;2:220. doi: 10.1016/s0140-6736(68)92656-1. [DOI] [PubMed] [Google Scholar]
- 4.Miller DT, et al. Consensus statement: chromosomal microarray is a first-tier clinical diagnostic test for individuals with developmental disabilities or congenital anomalies. Am. J. Hum. Genet. 2010;86:749–764. doi: 10.1016/j.ajhg.2010.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Friedman JM. High-resolution array genomic hybridization in prenatal diagnosis. Prenat. Diagn. 2009;29:20–28. doi: 10.1002/pd.2129. [DOI] [PubMed] [Google Scholar]
- 6.Brady PD, Vermeesch JR. Genomic microarrays: a technology overview. Prenat. Diagn. 2012;32:336–343. doi: 10.1002/pd.2933. [DOI] [PubMed] [Google Scholar]
- 7.Wapner RJ. A multicenter, prospective, masked comparison of chromosomal microarray with standard karyotyping for routine and high risk prenatal diagnosis. Am. J. Obstet. Gynecol. 2012;206:S2. [Google Scholar]
- 8.Hillman SC, et al. Additional information from array comparative genomic hybridization technology over conventional karyotyping in prenatal diagnosis: a systematic review and meta-analysis. Ultrasound Obstet. Gynecol. 2011;37:6–14. doi: 10.1002/uog.7754. [DOI] [PubMed] [Google Scholar]
- 9.Vetro A, et al. The introduction of arrays in prenatal diagnosis: a special challenge. Hum. Mutat. 2012;33:923–929. doi: 10.1002/humu.22050. [DOI] [PubMed] [Google Scholar]
- 10.Novelli A, et al. Microarray application in prenatal diagnosis: a position statement from the cytogenetics working group of the Italian Society of Human Genetics (SIGU), November 2011. Ultrasound Obstet. Gynecol. 2012;39:384–388. doi: 10.1002/uog.11092. [DOI] [PubMed] [Google Scholar]
- 11.American College of Obstetricians and Gynecologists. ACOG Committee Opinion No. 446: array comparative genomic hybridization in prenatal diagnosis. Obstet. Gynecol. 2009;114:1161–1163. doi: 10.1097/AOG.0b013e3181c33cad. [DOI] [PubMed] [Google Scholar]
- 12.Wapner RJ, et al. Integration of microarray technology into prenatal diagnosis: counselling issues generated during the NICHD clinical trial. Prenat. Diagn. 2012;32:396–400. doi: 10.1002/pd.3863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo YM, et al. Presence of fetal DNA in maternal plasma and serum. Lancet. 1997;350:485–487. doi: 10.1016/S0140-6736(97)02174-0. [DOI] [PubMed] [Google Scholar]
- 14.Devaney SA, et al. Noninvasive fetal sex determination using cell-free fetal DNA: a systematic review and meta-analysis. JAMA. 2011;306:627–636. doi: 10.1001/jama.2011.1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hill M, et al. Non-invasive prenatal determination of fetal sex: translating research into clinical practice. Clin. Genet. 2011;80:68–75. doi: 10.1111/j.1399-0004.2010.01533.x. [DOI] [PubMed] [Google Scholar]
- 16.Chan KC, et al. Hypermethylated RASSF1A in maternal plasma: a universal fetal DNA marker that improves the reliability of noninvasive prenatal diagnosis. Clin. Chem. 2006;52:2211–2218. doi: 10.1373/clinchem.2006.074997. [DOI] [PubMed] [Google Scholar]
- 17.White HE, et al. Evaluation of a novel assay for detection of the fetal marker RASSF1A: facilitating improved diagnostic reliability of noninvasive prenatal diagnosis. PLoS ONE. 2012;7:e45073. doi: 10.1371/journal.pone.0045073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kolialexi A, et al. Early non-invasive detection of fetal Y chromosome sequences in maternal plasma using multiplex PCR. Eur. J. Obstet. Gynecol. Reprod. Biol. 2012;161:34–37. doi: 10.1016/j.ejogrb.2011.12.025. [DOI] [PubMed] [Google Scholar]
- 19.Finning KM, et al. Prediction of fetal D status from maternal plasma: introduction of a new noninvasive fetal RHD genotyping service. Transfusion. 2002;42:1079–1085. doi: 10.1046/j.1537-2995.2002.00165.x. [DOI] [PubMed] [Google Scholar]
- 20.Clausen FB, et al. Report of the first nationally implemented clinical routine screening for fetal RHD in D-pregnant women to ascertain the requirement for antenatal RhD prophylaxis. Transfusion. 2012;52:752–758. doi: 10.1111/j.1537-2995.2011.03362.x. [DOI] [PubMed] [Google Scholar]
- 21.Scheffer PG, et al. Noninvasive fetal blood group genotyping of rhesus D, c, E and of K in alloimmunised pregnant women: evaluation of a 7-year clinical experience. Br. J. Obstet. Gynecol. 2011;118:1340–1348. doi: 10.1111/j.1471-0528.2011.03028.x. [DOI] [PubMed] [Google Scholar]
- 22.Traeger-Synodinos J, et al. Prenatal, noninvasive and preimplantation genetic diagnosis of inherited disorders: hemoglobinopathies. Expert Rev. Mol. Diagn. 2011;11:299–312. doi: 10.1586/erm.11.7. [DOI] [PubMed] [Google Scholar]
- 23.Scheffer PG, et al. Noninvasive fetal genotyping of human platelet antigen-1a. Br. J. Obstet. Gynecol. 2011;118:1392–1395. doi: 10.1111/j.1471-0528.2011.03039.x. [DOI] [PubMed] [Google Scholar]
- 24.Shammas C, et al. ThalassoChip, an array mutation and single nucleotide polymorphism detection tool for the diagnosis of beta-thalassaemia. Clin. Chem. Lab. Med. 2010;48:1713–1718. doi: 10.1515/CCLM.2010.331. [DOI] [PubMed] [Google Scholar]
- 25.Tsui NB, et al. Noninvasive prenatal diagnosis of hemophilia by microfluidics digital PCR analysis of maternal plasma DNA. Blood. 2011;117:3684–3691. doi: 10.1182/blood-2010-10-310789. [DOI] [PubMed] [Google Scholar]
- 26.Bianchi DW, et al. Fetal gender and aneuploidy detection using fetal cells in maternal blood: analysis of NIFTY I data. National Institute of Child Health and Development Fetal Cell Isolation Study. Prenat. Diagn. 2002;22:609–615. doi: 10.1002/pd.347. [DOI] [PubMed] [Google Scholar]
- 27.Fan HC, Quake SR. Detection of aneuploidy with digital polymerase chain reaction. Anal. Chem. 2007;79:7576–7579. doi: 10.1021/ac0709394. [DOI] [PubMed] [Google Scholar]
- 28.Lo YM, et al. Digital PCR for the molecular detection of fetal chromosomal aneuploidy. Proc. Natl. Acad. Sci. U.S.A. 2007;104:13116–13121. doi: 10.1073/pnas.0705765104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lun FM, et al. Microfluidics digital PCR reveals a higher than expected fraction of fetal DNA in maternal plasma. Clin. Chem. 2008;54:1664–1672. doi: 10.1373/clinchem.2008.111385. [DOI] [PubMed] [Google Scholar]
- 30.Chiu RW, et al. Non-invasive prenatal assessment of trisomy 21 by multiplexed maternal plasma DNA sequencing: large scale validity study. Br. Med. J. 2011;342:c7401. doi: 10.1136/bmj.c7401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehrich M, et al. Noninvasive detection of fetal trisomy 21 by sequencing of DNA in maternal blood: a study in a clinical setting. Am. J. Obstet. Gynecol. 2011;204:205, e201–e211. doi: 10.1016/j.ajog.2010.12.060. [DOI] [PubMed] [Google Scholar]
- 32.Palomaki GE, et al. DNA sequencing of maternal plasma to detect Down syndrome: an international clinical validation study. Genet. Med. 2011;13:913–920. doi: 10.1097/GIM.0b013e3182368a0e. [DOI] [PubMed] [Google Scholar]
- 33.Sehnert AJ, et al. Optimal detection of fetal chromosomal abnormalities by massively parallel DNA sequencing of cell-free fetal DNA from maternal blood. Clin. Chem. 2011;57:1042–1049. doi: 10.1373/clinchem.2011.165910. [DOI] [PubMed] [Google Scholar]
- 34.Sparks AB, et al. Noninvasive prenatal detection and selective analysis of cell-free DNA obtained from maternal blood: evaluation for trisomy 21 and trisomy 18. Am. J. Obstet. Gynecol. 2012;206:319, e311–e319. doi: 10.1016/j.ajog.2012.01.030. [DOI] [PubMed] [Google Scholar]
- 35.Bianchi DW, et al. Genome wide fetal aneuploidy detection by sequencing of maternal plasma DNA: diagnostic accuracy in a prospective, blinded, multicenter study. Am. J. Obstet. Gynecol. 2012;206:S367. [Google Scholar]
- 36.Chen EZ, et al. Noninvasive prenatal diagnosis of fetal trisomy 18 and trisomy 13 by maternal plasma DNA sequencing. PLoS ONE. 2011;6:e21791. doi: 10.1371/journal.pone.0021791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Palomaki GE, et al. DNA sequencing of maternal plasma reliably identifies trisomy 18 and trisomy 13 as well as Down syndrome: an international collaborative study. Genet. Med. 2012;14:296–305. doi: 10.1038/gim.2011.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sparks AB, et al. Selective analysis of cell-free DNA in maternal blood for evaluation of fetal trisomy. Prenat. Diagn. 2012;32:3–9. doi: 10.1002/pd.2922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ashoor G, et al. Chromosome-selective sequencing of maternal plasma cell-free DNA for first-trimster detection of trisomy 21 and trisomy 18. Am. J. Obstet. Gynecol. 2012;206:322, e321–e325. doi: 10.1016/j.ajog.2012.01.029. [DOI] [PubMed] [Google Scholar]
- 40.Norton ME, et al. Non-Invasive Chromosomal Evaluation (NICE) Study: results of a multicenter prospective cohort study for detection of fetal trisomy 21 and trisomy 18. Am. J. Obstet. Gynecol. 2012;207:137, e131–e138. doi: 10.1016/j.ajog.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 41.van den Oever JM, et al. Single molecule sequencing of free DNA from maternal plasma for noninvasive trisomy 21 detection. Clin. Chem. 2012;58:699–706. doi: 10.1373/clinchem.2011.174698. [DOI] [PubMed] [Google Scholar]
- 42.Ashoor G, et al. Fetal fraction in maternal plasma cell-free DNA at 11–13 weeks’ gestation: effect of maternal and fetal factors. Fetal Diagn. Ther. 2012;31:237–243. doi: 10.1159/000337373. [DOI] [PubMed] [Google Scholar]
- 43.Canick JA, et al. DNA sequencing of maternal plasma to identify Down syndrome and other trisomies in multiple gestations. Prenat. Diagn. 2012;32:730–734. doi: 10.1002/pd.3892. [DOI] [PubMed] [Google Scholar]
- 44.Benn P, et al. Prenatal Detection of Down Syndrome using Massively Parallel Sequencing (MPS): a rapid response statement from a committee on behalf of the Board of the International Society for Prenatal Diagnosis 24 October 2011. Prenat. Diagn. 2012;32:1–2. doi: 10.1002/pd.2919. [DOI] [PubMed] [Google Scholar]
- 45.Yao H, et al. Noninvasive prenatal genetic testing for fetal aneuploidy detects maternal trisomy X. Prenat. Diagn. 2012 doi: 10.1002/pd.3946. http://dx.doi.org/10/1002/pd.3946. [DOI] [PubMed] [Google Scholar]
- 46.Chiu RW, Lo YM. Noninvasive prenatal diagnosis empowered by high-throughput sequencing. Prenat. Diagn. 2012;32:401–406. doi: 10.1002/pd.3822. [DOI] [PubMed] [Google Scholar]
- 47.Lo YM, et al. Maternal plasma DNA sequencing reveals the genome-wide genetic and mutational profile of the fetus. Sci. Transl. Med. 2010;2:61ra91. doi: 10.1126/scitranslmed.3001720. [DOI] [PubMed] [Google Scholar]
- 48.Kitzman JO, et al. Noninvasive whole-genome sequencing of a human fetus. Sci. Transl. Med. 2012;4:137ra76. doi: 10.1126/scitranslmed.3004323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fan HC, et al. Non-invasive prenatal measurement of the fetal genome. Nature. 2012;487:320–324. doi: 10.1038/nature11251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fan HC, et al. Whole-genome molecular haplotyping of single cells. Nat. Biotechnol. 2011;29:51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lau TK, et al. Non-invasive prenatal screening of fetal sex chromosomal abnormalities: perspective of pregnant women. J. Matern. Fetal Neonatal Med. 2012 doi: 10.3109/14767058.2012.712569. http://dx.doi.org/10.3109/14767058.2012.712569. [DOI] [PubMed] [Google Scholar]
- 52.Bianchi DW. At-home fetal DNA gender testing: caveat emptor. Obstet. Gynecol. 2006;107:216–218. doi: 10.1097/01.AOG.0000199427.83503.d0. [DOI] [PubMed] [Google Scholar]
- 53.American College of Obstetricians and Gynecologists. ACOG Practice Bulletin No. 77: screening for fetal chromosomal abnormalities. Obstet. Gynecol. 2007;109:217–227. doi: 10.1097/00006250-200701000-00054. [DOI] [PubMed] [Google Scholar]
- 54.Snijders RJ, et al. UK multicentre project on assessment of risk of trisomy 21 by maternal age and fetal nuchal-translucency thickness at 10–14 weeks of gestation. Fetal Medicine Foundation First Trimester Screening Group. Lancet. 1998;352:343–346. doi: 10.1016/s0140-6736(97)11280-6. [DOI] [PubMed] [Google Scholar]
- 55.Wald NJ, et al. First and second trimester antenatal screening for Down’s syndrome: the results of the Serum, Urine and Ultrasound Screening Study (SURUSS) Health Technol. Assess. 2003;7:1–77. doi: 10.3310/hta7110. [DOI] [PubMed] [Google Scholar]
- 56.Malone FD, et al. First-trimester or second-trimester screening, or both, for Down’s syndrome. N. Engl. J. Med. 2005;353:2001–2011. doi: 10.1056/NEJMoa043693. [DOI] [PubMed] [Google Scholar]
- 57.Evans MI, Andriole S. Chorionic villus sampling and amniocentesis in 2008. Curr. Opin. Obstet. Gynecol. 2008;20:164–168. doi: 10.1097/GCO.0b013e3282f7321f. [DOI] [PubMed] [Google Scholar]
- 58.Shaffer LG, Bui TH. Molecular cytogenetic and rapid aneuploidy detection methods in prenatal diagnosis. Am. J. Med. Genet. C: Semin. Med. Genet. 2007;145C:87–98. doi: 10.1002/ajmg.c.30114. [DOI] [PubMed] [Google Scholar]
- 59.Cirigliano V, et al. Rapid prenatal diagnosis of common chromosome aneuploidies by QF-PCR, results of 9 years of clinical experience. Prenat. Diagn. 2009;29:40–49. doi: 10.1002/pd.2192. [DOI] [PubMed] [Google Scholar]
- 60.Boormans EM, et al. Comparison of multiplex ligation-dependent probe amplification and karyotyping in prenatal diagnosis. Obstet. Gynecol. 2010;115:297–303. doi: 10.1097/AOG.0b013e3181cbc652. [DOI] [PubMed] [Google Scholar]
- 61.Toutain J, et al. First-trimester prenatal diagnosis performed on pregnant women with fetal ultrasound abnormalities: the reliability of interphase fluorescence in situ hybridization (FISH) on mesenchymal core for the main aneuploidies. Eur. J. Obstet. Gynecol. Reprod. Biol. 2010;149:143–146. doi: 10.1016/j.ejogrb.2009.12.015. [DOI] [PubMed] [Google Scholar]
- 62.Hills A, et al. QF-PCR as a stand-alone test for prenatal samples: the first 2 years’ experience in the London region. Prenat. Diagn. 2010;30:509–517. doi: 10.1002/pd.2503. [DOI] [PubMed] [Google Scholar]