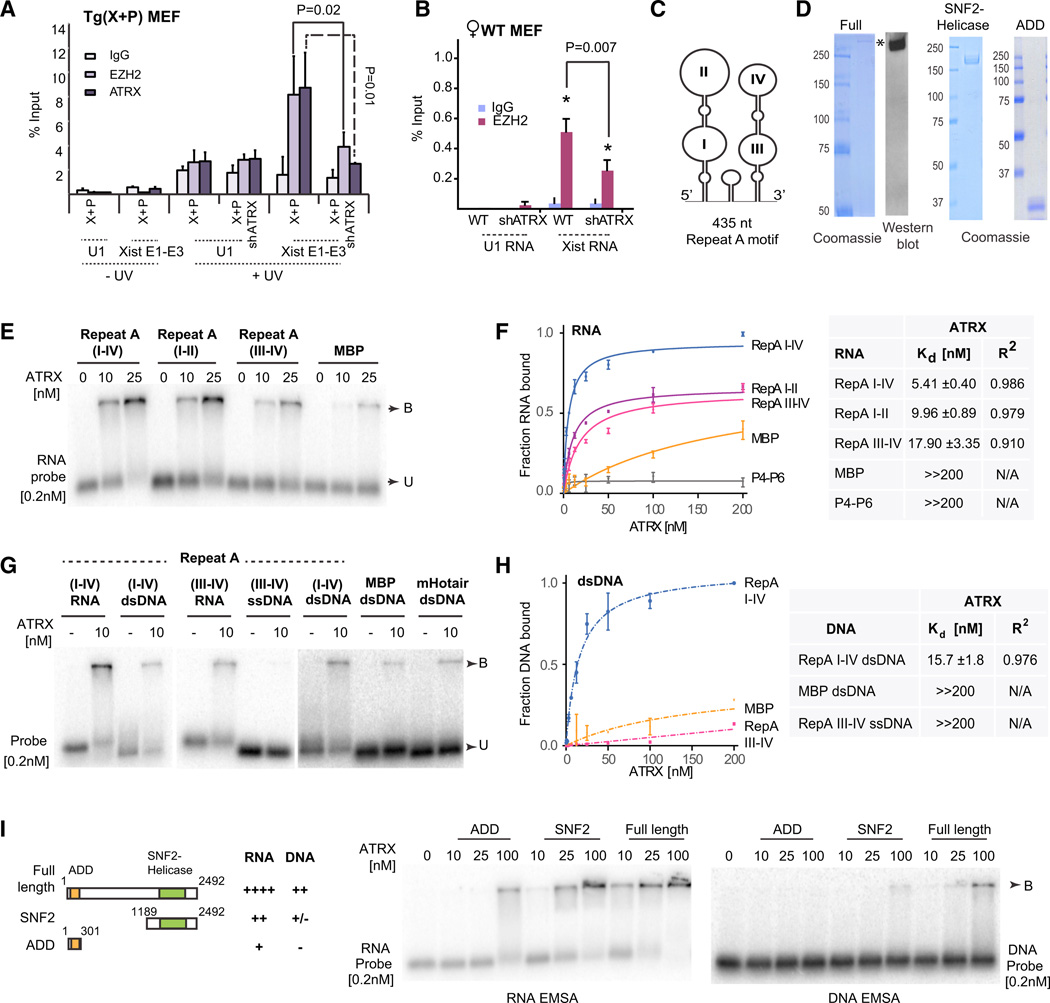
Figure 3. ATRX Binds RNA and Stimulates RNA Binding to PRC2.
(A,B) RIP ± UV crosslinking in X+P transgenic MEFs after dox-induction for 24 hr (A) or WTMEF(B) Primers spanning Xist exons 1–3 and U1 snRNAs were used for qPCR. One percent input was processed in parallel. SE from three independent experiments. P values calculated using Student’s t test.
(C) One possible structure of Repeat A (Maenner et al., 2010).
(D) Coomassie stain of purified full-length FLAG-ATRX-HA (left), C-terminal SNF2/helicase (middle), and N-terminal ADD (right) domains.
(E) RNA EMSA with ATRX at indicated concentrations and 0.2 nM of each probe. B, bound; U, unbound probe.
(F) Left: Binding Isotherms of ATRX for indicated RNAs. SE from three independent experiments shown. Right: table summarizing Kd’s and R2 values. ≫200 nM indicates Kd’s out of the assay range. N/A, not applicable.
(G) EMSA of ATRX with different RNA, dsDNA, or ssDNA probes.
(H) Left: Binding Isotherms of ATRX for indicated DNAs. SE for three independent experiments shown for each point. Right: table summarizing Kd’s and R2 values. ≫200 nM indicates Kd’s out of the assay range. N/A, not applicable.
(I) RNA and DNA EMSAs using ATRX truncation mutants, with summary of results.