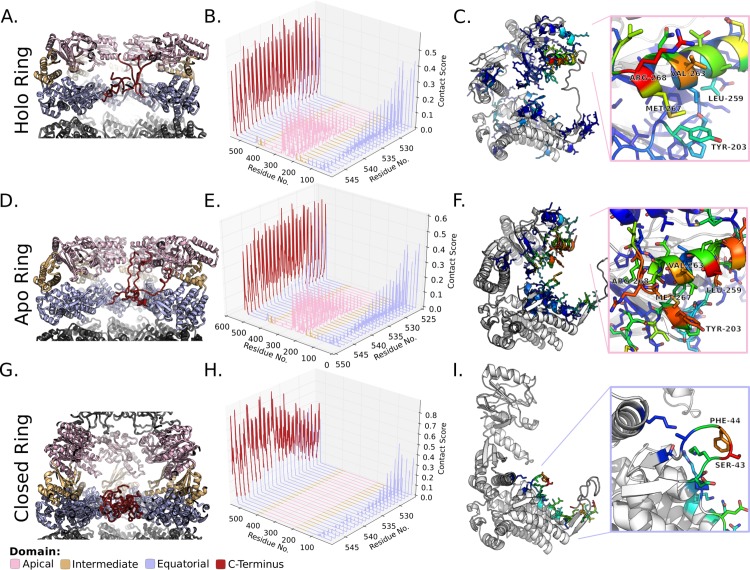
Fig 2. Characterization of C-terminal Contacts in GroE Simulations.
Residue contacts of the GroEL C-terminus from 190 ns of implicit solvent molecular dynamics simulations. A,D,G) Representative snapshots of the the GroE simulations colored by domain as indicated. B,E,H) Residue contacts between the terminal residues and the rest of the chaperonin residues from the open and closed state simulations. Higher values represent more frequent contacts. C,F,I) Heat maps of the terminal residue's (M548) contacts mapped onto representative monomer from the simulations. The monomer structures derive from the holo enzyme simulation (C), the trans-ring of the GroES bound (F), and the cis-ring of the GroES bound (I) simulations. The insets represent helix I (C,F) or the stem loop (I). C) A heat map of the holo ring's M548 contacts mapped onto a representative snapshot from the simulation. Inset: view of the apical domain helix I colored by M548 contact frequency. In panels C, F, & I contacts between M548 and other tail residues have been omitted by setting their values as zero. Structures were rendered in PyMOL.