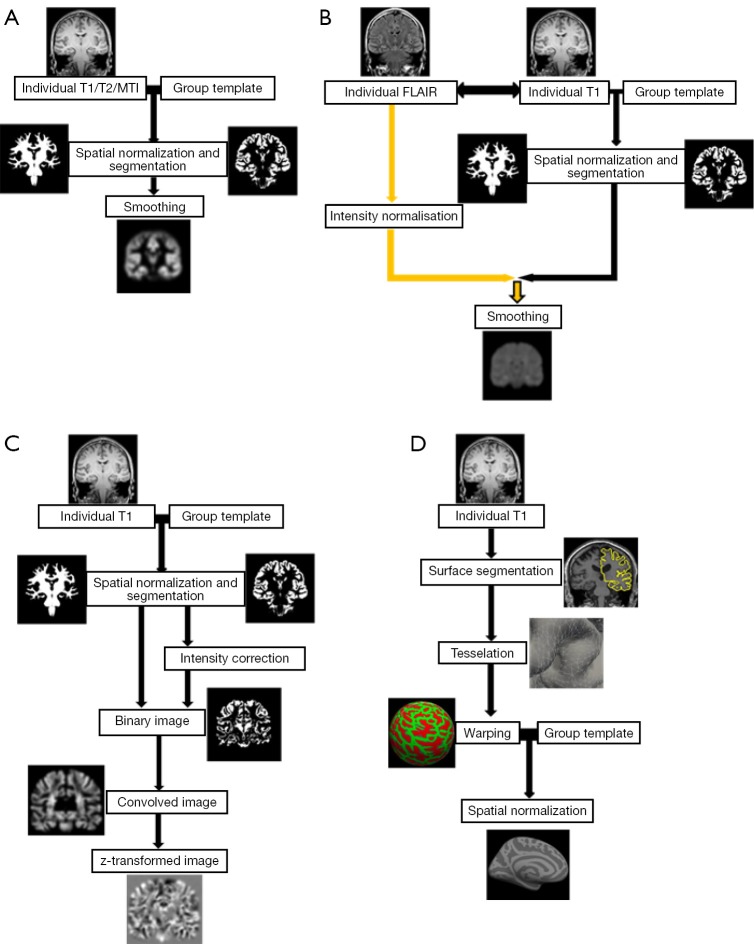
Figure 1.
Schematic overview of different VBM processing approaches. (A) Workflow of a “classical” VBM, e.g., in SPM, based on a T1-weighted image. The individual T1-weighted image is spatially normalized, segmented and smoothed. Similar workflows can be applied for magnetization transfer and T2-weighted images; (B) workflow for normalized T2-FLAIR signal intensity mapping. Segmentation and normalization is performed combined with a T1-weighted and T2-FLAIR image (after co-registered). As an additional processing step, intensity normalization of T2-FLAIR needs to be included. The other steps are similar to (A); (C) workflow following the MAP by Huppertz et al., exemplarily shown with the so called “junction map”. Normalization and segmentation are similar to classical VBM above including bias correction. Next, a binary image is created that represents the distribution of voxels with intensities between grey and white matter and then smoothed (convolved image). From the resulting image, the average of equally processed images of a normal population is subtracted and divided by the standard deviation to create a map of z-scores; (D) workflow of SBM, e.g., using Freesurfer, based on a T1-weighted image. As a first step, the boundaries between the gray/white matter and the pial surface are determined on the basis of a brain segmentation. Then the surfaces are tessellated by polygons whose meeting points or vertexes can be characterized by coordinates. To enable comparison between subjects, the brain images are warped into a common space, usually a sphere. On this basis the brains can be spatially normalized to a given template. VBM, voxel-based morphometry; SPM, statistical parametric mapping; MAP, Morphometric Analysis Programme; SBM, surface-based morphometry.