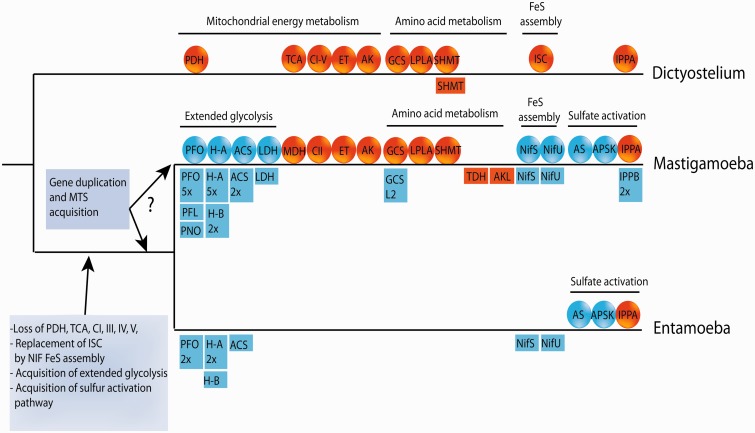
Fig. 6.
Evolution of selected biochemical pathways in amoebae. A common ancestor of mastigamoeba and entamoeba possibly lost majority of canonical mitochondrial enzymes/pathways such as PDH complex, majority of enzymes of TCA cycle, and respiration chain complexes with exception of CII. Mitochondrial ISC assembly was replaced by bacterial NIF machinery. Oxidative phosporylation was replaced by anaerobic substrate level phosporylation catalyzed by enzymes of extended glycolysis that were acquired by LGT. A common ancestor also acquired two components of sulfur activation pathway (AS and APSK). Gene duplication and acquisition of MTS resulted in dual localization of NIF system and enzymes of extended glycolysis in mastigamoeba. However, it is not clear whether these events happened in a common ancestor or only in mastigamoeba lineage. CI-V, respiratory complexes; ET, electron-transferring flavoprotein dehydrogenase (ETFDH), ETFa, and ETFb; GCS, glycin cleavage system; H-A/B, hydrogenase clade A/B; IPPA/B, inorganic pyrophosphatase clade A/B PFL, pyruvate formate lyase. Genes acquired by LGT are given in blue, genes of mitochondrial origin that were vertically inherited are highlighted by orange; squares and circles indicate cytosolic and mitochondrial localization, respectively; the numbers represent the number of cytosolic genes.