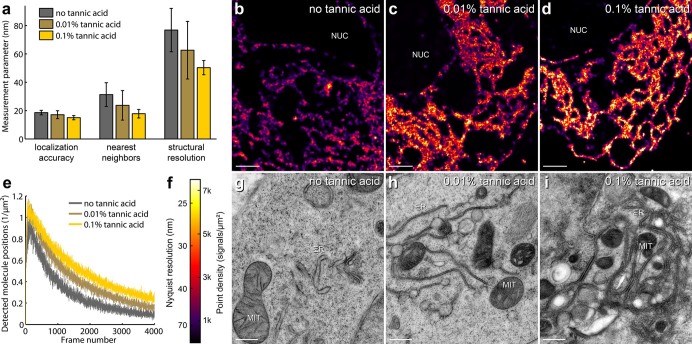
Figure 2. The quality of SMLM imaging is dependent on addition of tannic acid to the freeze substitution medium.
(a) Comparison of average single molecule localization accuracy, nearest neighbor distances and structural resolution of EphA2-mVenus localized in the plasma membrane and the endoplasmic reticulum (ER) of HEK293T cells in resin sections for samples with different concentrations of tannic acid in the freeze substitution medium. Nearest neighbor distances have been determined considering the 20 nearest molecule positions around each position. Error bars represent standard deviations. (b–d) Example SMLM images of cells, all acquired under the same conditions, depicting typical results for different concentrations of tannic acid. Scale bars are 1 μm. (e) Detected single molecule signals per μm2 and image frame during SMLM data acquisition for different concentrations of tannic acid in the freeze substitution medium. Only areas of fluorescently labeled structures were included. Each curve represents the average values of the SMLM measurements (total 57) for each concentration of tannic acid. (f) Color code for local density of detected molecules and Nyquist limited resolution in SMLM images (b–d). (g–i) Representative TEM images showing the typical quality of the ultrastructure (ER and mitochondria (MIT)) for freeze substitution with different concentrations of tannic acid. Scale bars are 0.5 μm. Note that the TEM images in (g–i) depict different cells than those shown in the SMLM images (b–d).