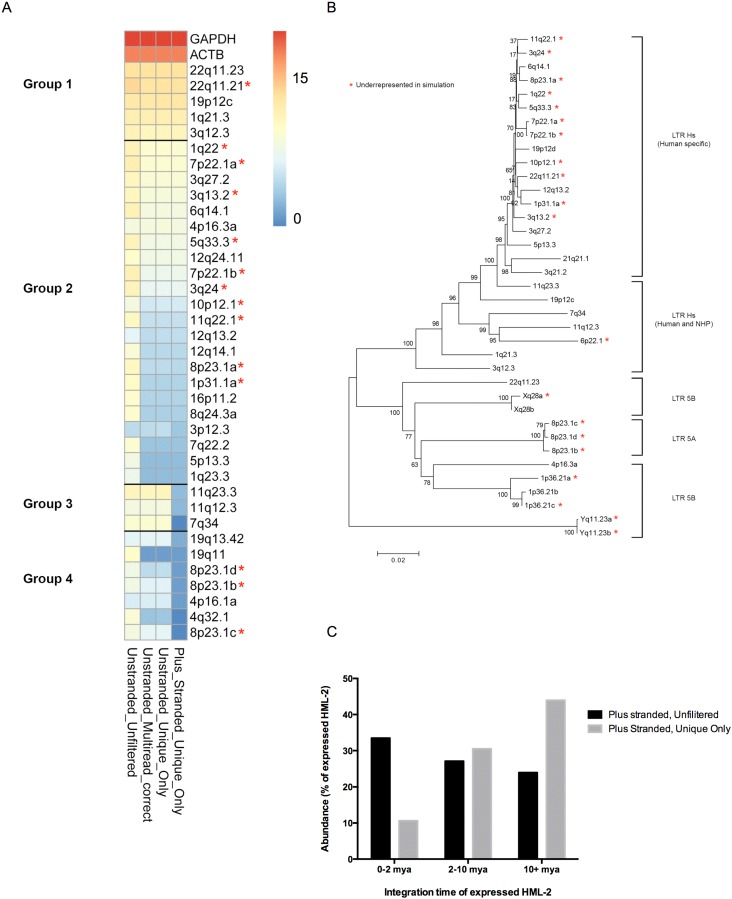
Figure 1.
RNASeq analysis of HML-2 expression in Tera-1 cells. (A) RNASeq reads derived from Tera-1 cellular RNA were aligned to the hg19 build of the human genome, using either a stranded (“Plus Stranded”) or unstranded (“Unstranded”) alignment. Aligned reads were either kept in full (“Unfiltered”), or were filtered based on mapping quality scores to only retain reads that uniquely aligned to one map location (“Unique Only”). The fragments per kilobase per million mapped reads (FPKM) values representing relative expression in Tera-1 cells were determined either with a multi-read correct parameter (“Multi-read Correct”) that proportionally allocates multi-reads to mapping locations, or without this parameter. FPKM values for selected HML-2 proviruses and the cellular genes GAPDH and β-actin (ACTB) across the analyses were log-normalized and used for heatmap generation to demonstrate the effects of the different analyses on expression levels. Proviruses and gene loci are divided into four groups according to their relative values following the different analyses: stable (Group 1); decrease after Unique Only (Group 2); decrease after Plus stranded alignment (Group 3); and decrease after Unique Only and Plus stranded analysis (Group 4). Log-normalized FPKM is shown by the colors from high (red) to low (blue), as indicated in the chart to the right. The (*) symbols refer to proviruses predicted to be underrepresented by 15% or more based on an in silico simulation. (B) A neighbor-joining tree of the underrepresented proviruses was created using the full provirus sequence. The p-distance method was used and bootstrap values are indicated as percent of 1000 replicates. (C) The abundance of transcripts after the Plus stranded, Unfiltered and the Plus Stranded, Unique Only analyses are plotted against estimated times of integration to show the effect of the Unique Only analysis on recently integrated proviruses. The 0–2 mya group includes human specific integrations with high sequence similarity predicted to be underrepresented in the Unique Only RNASeq in silico simulation. The relative abundance in Tera-1 cells was calculated for each provirus based on (provirus FPKM)/(total HML-2 provirus FPKM) × 100. Elements without 5’ or 3’ LTRs were unsuitable for age estimation and are not included.